Genomics 101: What is long-read sequencing?
By Florence Cornish onIn the series, ‘Genomics 101’, we go back to basics and explore some of the most important topics in genomics. In this blog, we explain what is meant by ‘long-read sequencing’, and how it could improve our approaches to cancer care.
First things first, what is DNA sequencing?
DNA is made up of 4 different ‘bases’, which we represent using the letters A, T, C and G.
To read the order of these letters, we use a technique called DNA sequencing.
The complete sequence of DNA in humans is called the human genome, and it’s over 3 billion letters long. It acts as the instruction manual for our bodies, allowing us to survive, function and grow.
When scientists sequence the genome, they must break it down into smaller fragments. This is because current technology cannot read the entire genome at once.
Scientists then piece these fragments back together to read the original sequence.
What is long-read sequencing?
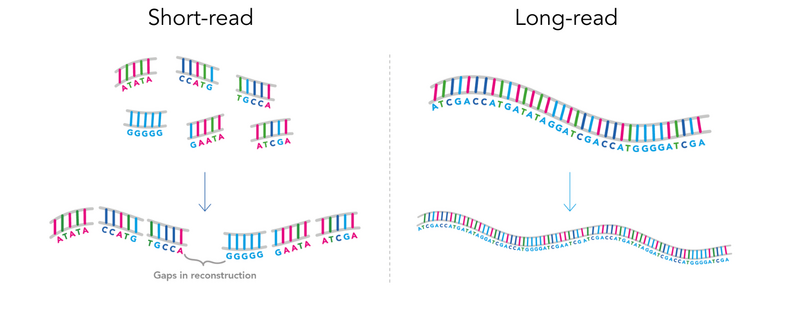
Long-read sequencing is a technique that lets us read much longer fragments of DNA than older sequencing methods.
In short-read sequencing, DNA must be broken down into small fragments of around 900 letters. For long-read sequencing, DNA is broken into much larger fragments, ranging from around 10,000 letters all the way to 100,000 letters in length.
Emma McCargow, Cancer Programme Lead at Genomics England, compares long-read sequencing to pages in a book:
“Imagine our genome as a library of books, in those books you have pages, and every page is a gene. In current sequencing methods, we read the genome by looking at those pages word by word. For long-read sequencing, we read the genome by sentences and paragraphs.”
What are the benefits and limitations of long-read sequencing?
Long-read sequencing is useful because it gives us large amounts of genetic information in a quick and effective way.
It can identify genetic changes that are not picked up by older sequencing methods and allows us to investigate sections of the genome that are relatively unexplored, sometimes referred to as ‘DNA dark matter’.
The information we get from long-read sequencing has the potential to transform diagnoses for genetic diseases such as cancer. This could ultimately lead to the development of new, personalised treatment pathways.
The downside of long-read sequencing is that it can have a lower accuracy per read than short-read sequencing. When both long-read and short-read techniques are paired together, they have the potential to make genetic information more accessible than ever before.
Long-read sequencing and cancer
Cancer is a disease of the genome. It occurs when changes in a person’s DNA result in cells growing and dividing uncontrollably.
These genetic changes can be inherited from a parent or occur later in a person’s life. They are often very difficult to spot.
Identifying these changes is extremely important. It helps clinicians to gain a clearer understanding of a patient’s diagnosis so they can recommend the best possible treatment.
Certain cancers are well-suited to long-read sequencing because they contain genetic changes that are difficult to find with traditional sequencing approaches.
An example of this is cancer caused by a genetic variation called a ‘repeat’, where sequences of letters are repeated several times in the genome.
Repeats are very difficult to detect using short-read sequencing, because they are often missed when the short DNA fragments are pieced back together. Long-read sequencing can detect these repeats much more effectively, allowing us to provide a rapid genetic diagnosis.
Long-read sequencing at Genomics England
Genomics England have been pioneers in bringing genomic technology to patients, and long-read sequencing is no exception.
Our Cancer 2.0 initiative aims to explore the best ways in which long-read sequencing, combined with other forms of data, could support quick and early diagnosis of cancer.
Long-read sequencing is already a very efficient technology. Therefore, our main focus is developing a pipeline that will allow the technology to be used in standard healthcare.
This pipeline would extend from collecting the sample from the patient (usually a simple blood test), through to sequencing the DNA, and ultimately using the information to make clinical decisions.
We are working closely with our partners at NHS England to explore how this process could enable long-read sequencing to be used in standard cancer diagnosis.
Want to find out more?
Find out more about our work on long-reads through the Cancer 2.0 initiative, or check out our video on the next generation of cancer sequencing for a more detailed explanation of the technology.
Prefer to listen? Catch our Genomics 101 podcast on long read sequencing.

