Genomics 101: What is whole genome sequencing?
By Florence Cornish onIn this series, ‘Genomics 101’, we go back to basics and explore some of the most important underlying topics in genomics. In this blog, we explain what is meant by ‘whole genome sequencing’, and how this technology is impacting healthcare.
First things first, what is a genome?
The human genome is the complete set of genetic material present in each person. It is the end-to-end sequence of DNA that acts as an instruction manual allowing our bodies to survive, function and grow.
This sequence is made up of 4 different ‘bases’, which we represent using the letters A, T, C and G.
The cells in our body can read this sequence of letters, almost like reading a book, and produce different types of proteins that we need to survive. The order of the letters in the sequence determines which protein is produced.
Each small stretch of letters that codes for a specific protein is known as a ‘gene’, and the human genome is made up of more than 20,000 of them.
Why is sequencing the genome important?
Our genome encodes all the vital information we need to survive.
If there are problems in the genome, for example letters in the wrong order, this can affect our health. For people with genetic conditions, looking at their genome can often help us find the cause of their condition and how we might help them.
There are lots of different ways that researchers can look at the human genome.
Older, more traditional methods let us look at the genome in small sections, for example one gene at a time. These methods are often time consuming and make it difficult to find the genetic cause of a patient’s condition because we must know exactly which genes to test, and they can only be looked at one at a time.
Since these older methods, there have been many great advances in sequencing. Faster, more automated techniques that sequence millions of short DNA fragments have emerged. We call these techniques ‘next generation sequencing’.
Examples of this include panel testing, in which hundreds of genes are sequenced at the same time, as well as whole genome sequencing, which is what this blog will focus on.
What is whole genome sequencing?
Whole genome sequencing is a technique that allows us look at someone’s entire genetic code. Where traditional methods just look at particular sections of the genome, whole genome sequencing aims to read the entire thing.
The first full human genome sequence was generated in 2003 under the Human Genome Project. It took over 10 years and a global network of researchers to do it. Since then, methods have become increasingly quick, and we can now sequence an entire genome in just one day.
How does whole genome sequencing work?
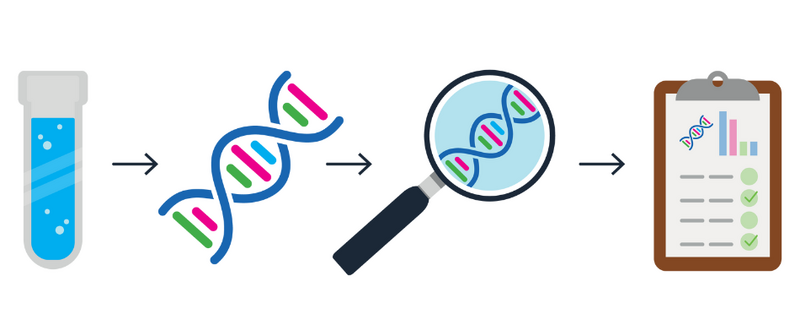
To sequence the genome, a healthcare professional must first take a sample from the patient. This can often be a simple blood test, as the DNA present in one drop of blood is enough for us to generate a sequence.
DNA is then extracted from the sample and ‘fragmented’, meaning it is broken up into smaller sections. These sections are then sequenced and put back together like pieces in a jigsaw, allowing us to read the entire sequence from start to finish.
Scientists and researchers can look at the genome sequence, searching for anything unusual that might be causing problems for the patient. They do this by comparing the patient’s genome to a ‘reference genome'.
By comparing all genomes to the reference genome, we can identify what is different in the patient’s genome that might be causing the problems.
What are the benefits and challenges of whole genome sequencing?
The main advantage of whole genome sequencing is that it can help us find a genetic diagnosis for patients. It can test a wide range of genes all at once, and detect several different types of genetic changes.
Because of this, whole genome sequencing can pick up genetic diagnoses that standard tests might have missed. It is the most comprehensive genomic test currently available, and is helping to bring answers to so many patients and families.
At Genomics England, we believe that whole genome sequencing will work best when used alongside other approaches. For example, it could be used alongside more traditional genetic tests, as well as other clinical information about the patient.
This will give us a clear, in-depth picture of a patient’s condition, and in turn the best chance of helping them.
However, a big difficulty for whole genome sequencing is storing all of the information it gives us. This can be challenging, given the huge amounts of data that are generated from sequencing an entire genome.
What can whole genome sequencing detect in cancer?
Whole genome sequencing can be very useful for treating patients with cancer.
To sequence the genome of someone with cancer, we must take the sample directly from the tumour as well as a standard sample from their blood.
This is because DNA in the tumour will be different from DNA in the non-cancerous cells, and we must compare the genome sequences between the 2 samples to see how they are different.
Once we have detailed genetic information about someone’s cancer, it may help us to find which treatment will work best, depending on their genomic changes. We call this ‘personalised medicine’, because we are choosing a treatment based off each individual.
Personalised medicine is still very much in its early stages, and it is not commonly carried out just yet. However, as whole genome sequencing becomes increasingly used, we could move away from a one size fits all approach to cancer therapy, and towards a specifically tailored treatment.
The 100,000 Genomes Project
The 100,000 Genomes Project was a landmark initiative that sequenced over 100,000 genomes from 85,000 participants between 2013 and 2018.
It was carried out by Genomics England in close partnership with the NHS, and aimed to uncover answers for participants whilst making whole genome sequencing a part of routine care.
Though sequencing for the project is now complete, approved researchers are looking through the DNA sequences every day to try and find answers for participants and their families.
The project led to the formation of the ‘National Genomic Research Library’ and the ‘NHS Genomic Medicine Service’, in which NHS patients can continue to have their genomes sequenced and stored, so that researchers can help to diagnose them.
In 2022 alone, over 1,500 new potential diagnoses were returned to clinical teams in the NHS thanks to research on data in the National Genomic Research Library.
What is the future of whole genome sequencing?
Many great advances over the past few years suggest a promising future for whole genome sequencing in healthcare.
As the technology becomes more efficient, we may continue to learn about the genome in more and more detail, and increase our understanding of genomics and disease.
We can expect large scale genomic data sets like the National Genomic Research Library to keep expanding as more people volunteer their data. We can also expect personalised medicine to become even more significant for treating genetic conditions.
These advancements provide a valuable new resource for the NHS, enabling us to continue bringing answers to patients and families and providing them with the best possible care.
To see how Genomics England will play a role in the promising future of genomics, check out our Research vision, as well as some of the work being done in our research community to help bring answers to patients.
Prefer to listen? Check out the Genomics 101 podcast on whole genome sequencing.

