The BioSample Journey
By John Pullinger and Tala Zainy onThe Genomics England Biosample Logistics team are responsible for monitoring the flow of samples and associated data submitted to the Genomic Medicine Service (GMS). In this blog, they will discuss the high-level journey of a sample in the GMS and in Research Cohorts.
As the Biosample Logistics Team at Genomics England, we have been dealing with patients and research participants since the beginning of the 100,000 Genomes Project.
This research project led to the creation of the Genomic Medicine Service (GMS), where we routinely monitor the flow of hundreds of samples submitted weekly across England from seven different Genomic Lab Hubs (GLHs).
Apart from the GMS, the wider Logistics Team also supports the monitoring of sample and genome flow for our various research collaborations. We ensure that biological samples and genomes progress through bioinformatics analysis successfully, adding further data into our National Genomic Research Library (NGRL) for controlled access to researchers and industry partners who work with us.
The Sample Journey: Genomic Medicine Service
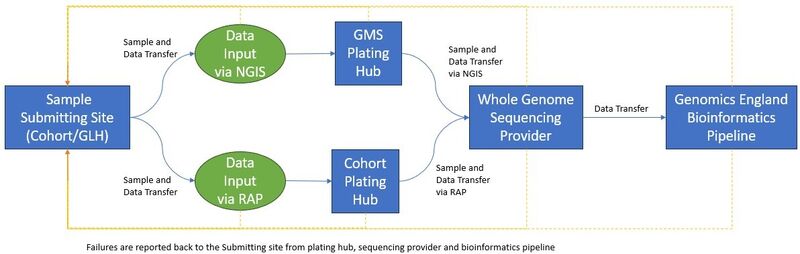
Samples for the GMS are initially sourced from NHS hospitals around England. They are extracted and prepared at 7 Genomic Laboratory Hubs (GLHs) which geographically cover the whole country. These GLHs send the samples onto a centralised Plating Hub, who are responsible for bringing the samples from different GLHs together into uniquely barcoded 96-well plates.
The physical samples are accompanied on their journey by a parallel data flow. This begins with the creation of a referral containing demographic, consent and basic level clinical data, i.e., clinical indication.
This data is then combined with sample level data and initial quality control information, such as sample volume and concentration, in the initial .csv file. This file is submitted by the GLH to our National Genomic Informatics System (NGIS).
The .csv file exchange continues through the biosample journey with checkpoints at the Plating Hub. This is where the samples which are physically received are compared to the list of samples which the GLH has marked as sent.
The samples are then prepared to be sent for sequencing, with related samples plated separately to reduce the risk of intra-referral contamination, as this is harder to decipher later in the bioinformatics pipeline.
Complexities in the pipeline
The role of Biosample Logistics is to monitor the end-to-end data and sample flow journey, and to facilitate the resolution of any file exchange or physical sample related issues. Examples of these complexities include samples at the Plating Hub with incorrect data, potential contamination, and missing samples within a set consignment.
Once the samples have arrived at the sequencing facility, they are immediately visually checked to ensure there is adequate volume in each well for sequencing with a Pass/Fail status. This is reported back via another .csv file, and any discrepancies between physical plate and data are flagged to Genomics England.
The samples then have their concentration measured using PicoGreen®, which is a fluorescent nucleic acid stain for quantitating double-stranded DNA. This is then reported back to Genomics England.
All passing samples continue onto library preparation and sequencing processes, and those sequences that pass the sequencing provider’s analytical and contractual metrics are returned to the Genomics England Bioinformatics pipeline for automated analysis and reporting of results on the Interpretation Portal.
Data from the file exchange is visually presented in a sample tracking tool, which has multiple tabs to split the data into each stage of the sample’s journey, as well as overall case flow tabs. In addition to this, a key part of the pipeline is failure reporting:
- Data Quality Report – covers data issues that affect the submission of the case to bioinformatics, and discrepancies between clinical data submitted via the test referral and sample data submitted via csv. With the exceptions of sensitive data queries, these data failures are usually amendable through data corrections via queries with GLHs, rather than sample replacements.
- Sample Failure Report – contains a list of failures for the physical samples such as an out-of-range concentration, potential sample swaps and contamination failures, as well as sequence data related failures (based on metrics agreed between GEL bioinformatics and the whole genome sequencing provider). This provides a view for these failures at sequencing provider and through the Genomics England bioinformatics pipeline. This allows the GLH to monitor, investigate and replace samples as required live throughout the clinical service. To note, the failure rate in the pipeline is low, and the majority of the samples pass first time around successfully.
Members of the Biosample Logistics Team are involved in the production and maintenance of specialised trackers and use of visual analytic platforms to monitor trends, case/sample level tracking, and process improvement suggestions with other stakeholders.
The Sample Journey – Research Cohorts
Research cohorts are sent a bulk upload file, which is a template that consists of multiple tabs to gather mandatory and optional data (minimum data requirements).
These include participant (gender, ethnicity etc.) and sample information (concentration, volume, sample identifiers etc.). The bulk upload is designed to capture all the required information for all downstream processes.
A guidance document is provided for research cohorts alongside the bulk upload to indicate the valid enumerations for the fields. Once completed, the bulk upload is emailed to the Cohort Manager at Genomics England for QC by a Biosample Operations Manager and Data Manager to validate that data meets expected requirements.
Updating the process
The Data Manager is responsible for creating the cohort study to allow for sample submissions. Previously, this process involved taking the bulk upload and creating .XML files per submission, either manually or via the use of a script. This was a time-consuming task (a minimum of half a day per submission), often requiring additional support from the Biosample Operations Managers.
In turn, this would delay the shipment of samples, as they could not be submitted until data was successfully ingested into the cohort sample tracking portal called RAP (research acquisition processing portal).
The need for a more efficient system was recognised and built by the product squad responsible for the sample tracking portal.
The system could accommodate direct ingestion of the bulk upload file as it is, without any intervention. The process now takes less than a minute per upload. If there are any errors in the upload, they are specified in the message with guidance on how to resolve.
Monitoring exchange and failure reporting
Biosample Operations managers are responsible for the monitoring and file exchange between the Plating Hub and the Sequencing Provider.
Research cohort samples can go directly from the research cohort submitting site to the sequencing provider. For example, long-read sequencing at Genomics England’s internal Research & Development (R&D) laboratory.
Alternatively, the samples are submitted (usually in batches) to the biorepository. This is for further QC checks and normalising into uniquely barcoded plates for short-read sequencing, or reformatting into 2D barcoded tubes for long-read sequencing.
Once samples are at the sequencing facility, further QC is carried out before proceeding to sequencing. If samples fail to meet the sequencing criteria, an attempt to send a repeat sample is made (if repeats have been agreed with the research cohort or if sufficient repeat sample is available).
Failures are reported via 3 methods by sequencing provider: visual QC fail, quantification fail (pre sequencing), and sequencing fails; such as sufficient quantity for library preparation (sequencing step) or on contractual metrics (post sequencing).
Failure reporting is generally done per batch, as they are processed and reported via email to Genomics England. After sequencing, the genomes are delivered to Genomics England to be run through the bioinformatics pipeline.
Where next?
Over the years, improvements have been made to several of the existing processes. For example, the introduction of lower quantification thresholds for acceptance of samples at the sequencing facility.
Any new changes to the pipeline require validation and monitoring over time. The Biosample Logistics Team is responsible for monitoring new changes and reviewing data before and after a given change. We aim to work with the wider internal and external stakeholders to reduce inefficiencies within the sample operational flow; for example, monitoring any trends in failures, and implementing changes to reduce these.
How are we improving the sample flow?
We are making a great effort to look at alternative methods for library preparation to reduce the hands-on time at the sequencing facility. This would reduce the overall turnaround times for patients/research participants.
Additionally, we are striving to make further improvements with key suppliers. The recent introduction of new sample pathways widen the initial source material types that can be submitted for sequencing without compromising the genome sequencing quality, and equally reducing the need for invasive sample collection methods.
Work is ongoing to test out new sample types for cancer such as fluid tumours (e.g., DNA from ascites for ovarian cancer, and pleural fluid for lung). Work is also underway to build a “tumour first” pipeline, which will allow tumours to be processed initially without the need for a matching germline (with the intention being that a germline sample can be submitted and analysed with the tumour at a later stage).
All of these innovations require a close working relationship with the internal product management teams for any technical changes, as well as alignment on changes with the wider stakeholders (GLHs/plating hubs/sequencing provider etc.)
Changes to the overall pipeline
One of the current inefficiencies in the overall sample flow is a given sample being quality checked (QC) at multiple stages. An improvement for the future would be to co-locate the end-to-end DNA extraction to genome processing pipeline to one facility, instead of multiple sites. This would minimise QC variation, reduce quality step redundancies, and improve the turnaround time from sample submission to results.
Data exchange systems between the various stakeholders in the pipeline could also be improved so that users could be notified where action needs to be taken. This differs from the current process, which requires users to log into the Genomics England systems and monitor issues live.
In short, our team will continue to focus our future efforts on working with our various stakeholders to optimise the sample flow process. We are working to widen the net for samples submitted via GMS and research cohorts, to provide maximal benefit to patients and researchers alike.
To see more updates from the bioinformatics team, check out our other blogs.


