100,000 Genomes Project
Genomics England's very first initiative – sequencing 100,000 genomes from around 85,000 NHS patients affected by rare disease or cancer – is leading to groundbreaking insights and continued findings into the role genomics can play in healthcare.

The impact of the project
The 100,000 Genomes Project was a British initiative to sequence and study the role our genes play in health and disease. Recruitment was completed in December 2018, although research and analysis is still ongoing.
Our participants have already helped us find actionable results for many patients with rare diseases and cancer.
18.5%
of data from the Project turned into actionable findings
85K+
participants' genomes sequenced for the Project
100K+
genomes sequenced by December 2018
Aims of the 100,000 Genomes Project
Make genomics part of routine healthcare
by working closely with the NHS to integrate whole genome sequencing
Enhance genomic healthcare research
by creating the largest genomic healthcare data resource in the world
Uncover answers for participants
both now and in the future through genomic-level analysis of conditions
Demonstrating impact
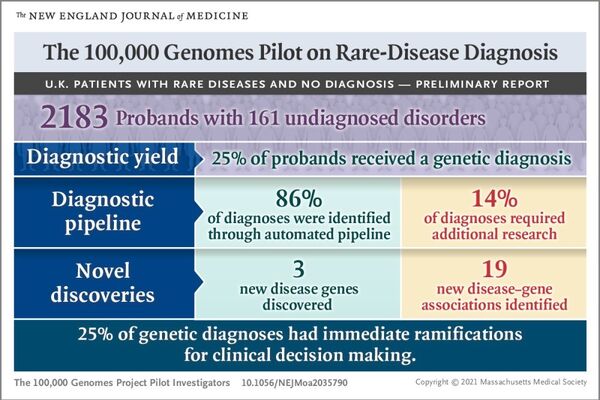
A world-first study using data on the 100,000 Genomes Project, and published in the New England Journal of Medicine, has demonstrated for the first time that whole genome sequencing (WGS) can uncover new diagnoses for people across the broadest range of rare diseases investigated to date and could deliver enormous benefits across the NHS.
The study looked at over four thousand people from over two thousand families who were early participants in the 100,000 Genomes Project. It found that using WGS led to a new diagnosis for 25% of the participants. Of these new diagnoses, 14% found variations in regions of the genome that would be missed by other methods, including other types of non-whole genomic tests.
Additional findings analysis
Our additional findings analysis focuses on looking for gene alterations in a specific list of genes that may increase the risk of people developing certain health conditions. In the case of increased chance of disease, steps can be taken to reduce the likelihood of the associated health condition developing or the condition can be treated and monitored.

Check Your Choice portal
Participants have the opportunity to check whether they have consented to receiving additional findings results, and to let us know if they've changed their minds.
Future of the project
Results from the 100,000 Genomes Project are still being returned to participants.
Beyond direct results, an additional and critically important spin off is the importance of this huge amount of data to researchers. Researchers are still using the data from the Project (and will continue to do so for many years to come) to develop new treatments, diagnostics, devices and medicines.
Only data that participants have consented to share is available to these approved researchers for further study.
Explore other initiatives
Diverse Data
Working in collaboration with researchers and the NHS, our Diverse Data…
Cancer 2.0
Our initiative to integrate long-read sequencing technology and multimodal…
COVID-19 Study
Read about our partnership with the GenOMICC consortium, led by the…