Learning meaningful data representations for biomedical research
By Francisco Azuaje on
In this blog post, Francisco Azuaje, Director of Bioinformatics at Genomics England, explains the concept of data ‘embeddings’ and how they are helping to advance biomedical research.
Data embeddings sit at the centre of cutting-edge machine learning applications for research that uses several types of data.
Machine learning continues to enable important progress across several areas of biomedical research and healthcare. This, in turn, is improving the way researchers and clinicians detect and treat disease using different kinds of data, e.g., genomics, other -omics, imaging and natural language.
This is accomplished using techniques based on neural networks that automatically ‘learn’ predictive features from relatively unstructured, ‘raw’ datasets.
Such advances provide a basis for bringing benefits to patients in the context of different applications, ranging from diagnosis and prognosis to predicting treatment responses. This requires the processing of massive, noisy and high-dimensional data, which is used as input for a variety of machine learning models.
Strengthening the trust of users requires us to demonstrate the validity (including predictive performance) and the utility (including estimates of the risk-benefit trade-off for patients) of these models.
In some cases, this will result in a better understanding of the biological mechanisms that underlie a prediction or medical condition. The latter is often possible by characterising the statistical associations between predictions, internal model parameters, and biological information not included in the model.
Learning embeddings: Rationale and definition
Despite a great diversity of techniques, there is a basic connection among them: they transform their inputs into compressed representations of numerical values. These features will, in turn, undergo further transformations depending on which specific application is under consideration.
Such transformations (e.g., converting sequences of letters to numbers, or going from very large images to compact numerical representations) bring challenges and opportunities.
A key challenge is preserving the patterns encoded in the original data, including an effective separation of signal from noise. The opportunities are abundant, such as the capacity to implement faster and more powerful models for making predictions about the data.
Therefore, we need to find effective ways to represent sequences of text (including genomes), images, or other data modalities with highly informative sets of numbers. The latter is referred to as embeddings, a concept which underpins some of the most promising advances in applied machine learning for biomedical research and healthcare.
The concept of embeddings may also be interpreted in the context of data dimensionality reduction, i.e., reducing the number of features held within a dataset.
However, unlike traditional dimensionality reduction techniques, such as PCA and SVD, machine learning-based techniques do not make specific assumptions about the underlying structure of the data. This includes linearity assumptions, or the type of patterns searched. Thus, machine learning-based embedding generation is particularly applicable to large and complex datasets.
To illustrate the conversion of input data into embeddings, Figure 1 shows examples for 3 data types that are commonly used to support biomedical research and healthcare applications.
Here, we have the case of DNA sequences consisting of symbols (A, G, T, C), that are transformed into 2 numerical values per sequence (1A). We also have images associated with tissues (e.g., tumour samples with different responses to a treatment), that are converted to a vector of 4 numbers (1B). Finally, we have natural language text describing different medical conditions, whose embeddings contain 3 values (1C).
In practice, the lengths of the inputs and embeddings are much larger, and vary according to the characteristics of the data and model selected for generating the embeddings.
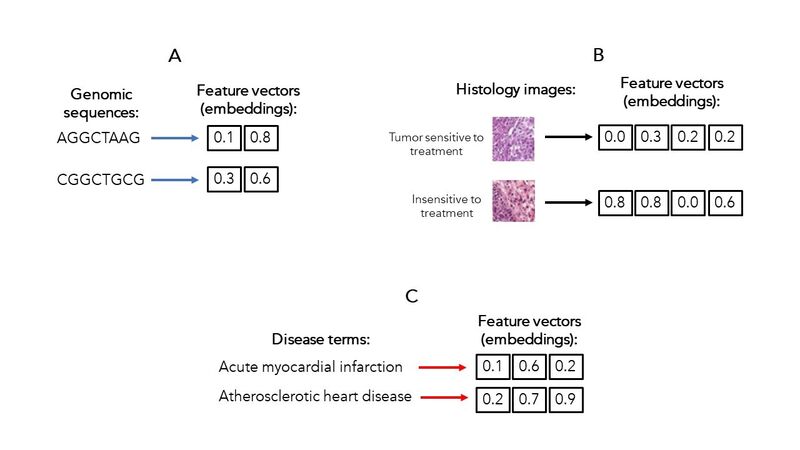
Figure 1. Example embeddings for different data modalities typically used in biomedical research and healthcare. Hypothetical examples of data inputs and their corresponding embeddings: A. For DNA sequences (8 letters transformed to 2 numerical values per sequence); B. For histopathology images (from hundreds of pixels to 4 numerical values per image); and C. For disease terms (from text with different meanings to 3 numerical values per clinical term).
A deeper look into the meaning and properties
A good embedding provides a compact (i.e., lower dimensional) representation of the original input, while synthesising relevant information about the structure or even meaning of the data.
For example, Figure 2 visualises multiple DNA sequences represented by 2-dimensional embeddings (X, Y). In this case, the embeddings encode enough information to account for the differences (and similarities) among the sequences based on 3 biological roles.
Generally, these plots are useful for machine learning practitioners and other users to visualise the results. In practice, 2- or 3-D visualisations are data projections produced by additional feature vector reduction techniques, and the biological meaning of the embeddings is not known in advance.
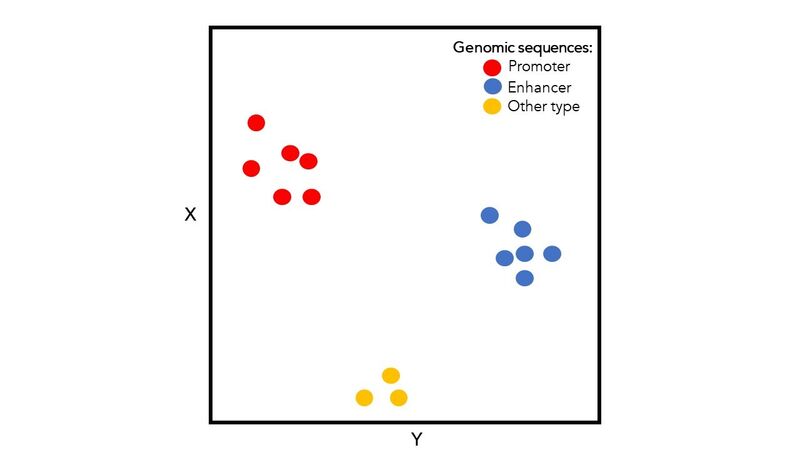
Figure 2. Visualization of embeddings on a 2-D plot. Examples of embeddings generated for 15 genomic sequences, associated with 3 groups of sequences with distinct biological roles. For illustration, we focus here on 2-dimensional embeddings with values displayed on the horizontal (Y) and vertical (X) axes.
There are other mathematical properties that are linked to the idea of a good embedding. For instance, embeddings are expected to disentangle the most important underlying sources of variation in the data. They also reflect the hierarchical organisation that is encoded in complex data sets.
This means, for example in the case of images, extracting key morphological features as well as more abstract features related to image content at multiple resolutions.
One could argue that the best embeddings are those that sufficiently approximate the processes underpinning the system of interest, e.g., the biology underlying the data.
Another key requirement is that the embeddings should represent information that is relevant for different applications. For example, embeddings that capture enough information for enabling image classification, can also be useful for enabling image segmentation.
In general, what makes an embedding a good data representation will depend on the intended ‘downstream’ uses. This involves different approaches to establishing the biomedical validity or relevance of the embeddings. Furthermore, embeddings greatly facilitate the combination of multiple data sources for different applications, including prediction models and similarity-based search.
So, now we have covered why embeddings are needed and what they represent, we may move on to how embeddings are obtained with different data types.
Using neural networks for embedding-based applications
Over the past decade, crucial advances in the field have largely been fuelled by a variety of neural network models. These techniques have allowed both power and flexibility to generate embeddings for multiple data modalities and their integration. They include many approaches with names like ConvNets, GNNs, and Transformers.
These neural networks differ in their original applications for specific data types, e.g., ConvNets, GNNs and Transformers for images, massive graphs, and text sequences respectively. They also differ in the way that transformations are applied to their inputs.
Regardless of input-types and algorithms used for generating embeddings, the same fundamental steps are typically required. Figure 3 portrays a simplified overview of this process using a DNA sequence as input to a “general purpose” neural network.
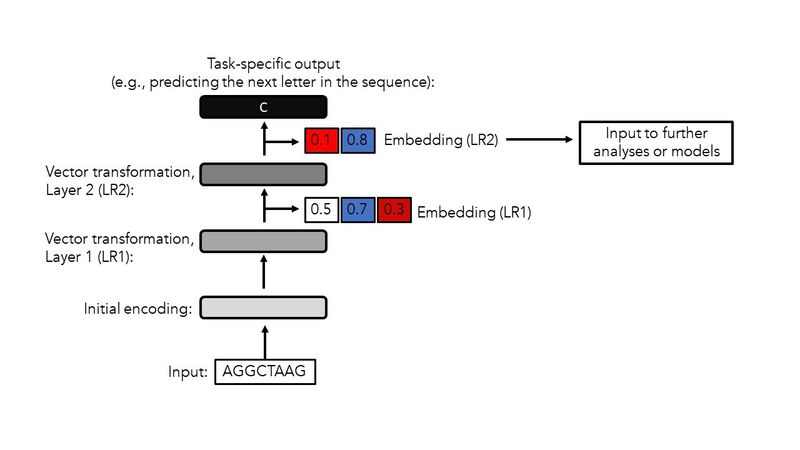
Figure 3. A simplified overview of key steps for generating embeddings. This example uses a DNA sequence as the input. The same general workflow is applicable to other types of data. LR: Layer.
Once the input has been presented to neural network, an initial transformation of the data is completed. This often involves a relatively simple encoding of the data, e.g., a one-hot encoding, which converts each text symbol into a binary representation.
Initial encodings can also be obtained by applying other neural networks that have been pre-trained with other datasets. For example, a network that has been trained to classify non-medical images and extract features associated with shapes and textures in these images.
The initial encoding is followed by multiple layers of linear and non-linear transformations. These transformations are computed with mathematical expressions that often consist of millions or even billions of parameters. Each layer can also contain multiple modules (or sub-layers) of transformations. Thus, the outputs of each layer represent different versions of embeddings, each capturing distinct aspects of the data structure and content.
In Figure 3, the first and second layers (LRs) generate embeddings consisting of 3 and 2 numerical values respectively.
The embeddings will be further processed and examined to assess their validity and applicability in downstream applications. In our example, this may include the classification of sequences in terms of functional classes (as in Figure 2) using other models, including but not limited to other neural networks.
Deeper into the algorithms: How are networks trained?
So far, we have assumed the network as already trained, i.e., the mathematical parameters of the different layers have been already estimated and can be applied to the incoming sequences. So, how does this happen?
Network parameters are adjusted via an optimisation algorithm that guides the network to implement a pattern recognition task. This involves many iterations of output generation, error estimation, and parameter adaptation. To compute errors, the true outputs for a given input are already available in the data, i.e., the network ‘self-learns’ the task.
In Figure 3, the task chosen is the prediction of the next letter for a given sequence (e.g., correctly predicting the letter ‘C’ for the putative input ‘AGGCTAAG’). This process of input presentation and parameter optimization is repeated multiple times for thousands or even millions of inputs.
The training process can be implemented in different ways. An alternative approach could be, e.g., for a given input: learning to recognise randomly selected sub-sequences that are masked during the training process.
A self-learning strategy has many advantages. It is sufficiently flexible for achieving different pattern recognition tasks, and specific enough to capture key properties embedded in the inputs. Moreover, it does not require the annotation (or labelling) of inputs by human experts, which is time consuming and often unreliable.
Further thoughts?
Here we shall conclude this brief introduction to the concept of data embeddings, why they are needed, and how they are usually obtained. Keep the conversation going and share your thoughts with us in the comments below.
Stay tuned for more exciting updates about the ongoing work at Genomics England. In the meantime, catch up on other posts from our series on bioinformatics and data science here.
About the author
Francisco Azuaje is Director of Bioinformatics at Genomics England and leads the applied machine learning team. He brings research and leadership experience from academia and the private sector, including the pharmaceutical industry, and is an Honorary Fellow of The University of Edinburgh. He has led research teams responsible for the analysis and predictive modelling of preclinical and clinically oriented datasets, including multi-omics, imaging data and natural language.
Acknowledgements
Thanks to the applied machine learning chapter (A. Rogerio, S. Ng, S. Barnett, T. Dyer), L. Valis, R. Sullivan, M. Mendonca and A. Rendon for their helpful feedback, and M. Blanksby and F. Cornish for editing and communications support.


