A decade of PanelApp: Exploring Genomics England's gene-disease knowledge base
By Eleanor Williams onPanelApp, Genomics England’s gene-disease database, has reached its 10th anniversary! Let’s look at how it has evolved over the past decade and the incredible impact it has had for genomic interpretation.
What is PanelApp?
PanelApp is a public knowledge base of genes that are linked to rare conditions and cancer susceptibilities. The data can be viewed, searched and downloaded through the PanelApp website and the Application Programming Interface (API).
Why is PanelApp different to other gene-disease databases?
In PanelApp, the gene-disease associations are grouped into lists of genes linked to similar conditions, which we call panels. This grouping is not done in other widely used gene-condition resources, such as ClinGen and OMIM.
The gene panels in PanelApp can be used to prioritise genomic changes in genes that are linked to conditions similar to those of a patient. This increases the likelihood of identifying the causal variant, while reducing the chance of incidental findings (a finding that is not related to the purpose of the test). For example, for a patient with bone abnormalities, the ‘Skeletal dysplasia’ panel can be used to prioritise genomic changes for clinical scientists to investigate further.
A unique feature of PanelApp is that clinical experts from the NHS in the UK and around the world can add and review gene-disease evidence. This crowdsourcing of knowledge has proven invaluable in keeping the gene panels up to date, and PanelApp has several hundred active expert reviewers who have left thousands of reviews on gene panels.
What has changed in the last 10 years?
The knowledge base has grown considerably over the last 10 years. It has gone from being used for research in the 100,000 Genomes Project, to now being part of the NHS England Genomic Medicine Service.
In September 2015, the first 15 gene panels in PanelApp were released for use. The simple evidence rating system developed (green/amber/red, with green being the highest ‘diagnostic’ level of evidence), along with crowdsourcing of knowledge. This crowdsourcing included gene lists gratefully received from UK Genetic Testing Network, Radboud University Medical Center, Emory Genetic Laboratory and Illumina, and meant that curators could rapidly assess the strength of published gene-disease evidence and build a comprehensive dataset. By the end of the recruitment and sequencing phase of the 100,000 Genomics Project in 2018, there were 175 gene panels.
Today, there are over 400 publicly available gene panels in PanelApp. From these, 302 are used in the NHS Genomic Medicine Service. Some of these panels are used in whole genome sequencing analyses performed by Genomics England, and others document the genes for more targeted tests carried out in the Genomic Laboratory Hubs in England. PanelApp has become a single searchable resource for all rare and inherited disease gene panel tests in the Genomic Medicine Service.
The number of genes linked to rare conditions continues to grow; since 2022 the number of new genes on the NHS Genomic Medicine Service panels with sufficient evidence for use in genomic analyses has increased by nearly 900, and since a gene may appear on more than one panel, over 4000 green-rated gene-disease relationships on the Genomic Medicine panels have been added, helping to diagnose many different conditions.
The impact of PanelApp
Patient outcomes
In the past 10 years, over 120,000 rare disease and cancer cases have been analysed at Genomics England using gene panels from PanelApp, with approximately 25% of rare disease patients receiving a diagnosis. Gene panels are integral to genomic testing in the Genomic Medicine Service, with green rated genes directly leading to the genomic diagnoses and improved care for patients. Since new knowledge can lead to more diagnoses, it is vital to keep the gene panels up to date.
Feedback of scientific discoveries into patient care
Patient data consented for research in the National Genomic Research Library has led to the discovery of novel gene-disease associations. Once published in a peer reviewed journal, the PanelApp gene panels can be updated for the benefit of future patients, enabling the completion of the feedback loop. An impactful example of this is the discovery of the RNU4-2 non-coding gene and its association with neurodevelopmental disorders, which is now green rated on 5 GMS panels listed in PanelApp.
Worldwide impact
A 2019 publication by Rueda-Martin et al described the PanelApp knowledgebase, and the paper has since had over 300 citations by other scientific publications according to EuropePMC. From this we can see that the gene panels have been used in the analysis of many patients’ genomes around the world. The gene panel data has also been added to other resources such as the UCSC Genome Browser and Open Targets, and therefore reaches a wide scientific audience.
Another way to look at the use of PanelApp is by the number of requests for the data. In June 2025, the PanelApp website and API had approximately 5 million requests from 1.8 million visits (each visit can make several requests). Requests come from around the world as shown in Figure 1.
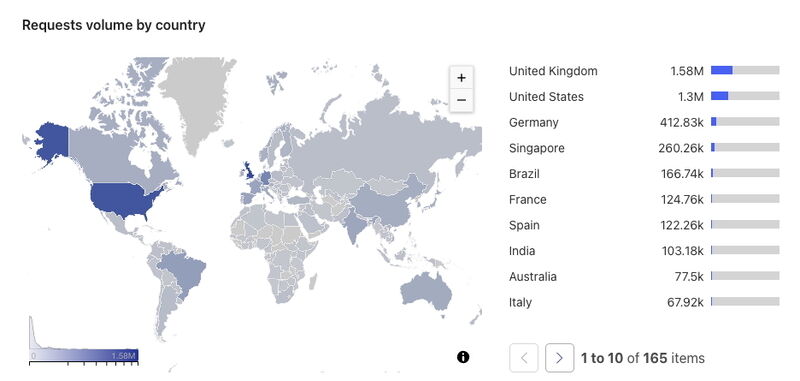
Figure 1 World map showing the requests to PanelApp by country and listing the top 10 countries that requests come from.
One of the most exciting developments for PanelApp in the past 10 years has come from the open sourcing of its code via GitLab. Australian Genomics has used this to build their own gene-disease knowledgebase, PanelApp Australia, with gene panels tailored to their own genomic services. A strong collaboration has developed between the 2 initiatives, which has facilitated both the exchange of information described in Stark et al, 2021 and skills, helping to keep the panels up to date for both services.
What’s next for PanelApp?
PanelApp gene panels remain a vital part of the NHS England Genomic Medicine Service with a crucial collaboration between Genomics England and NHS England enabling continued updates.
We also continue to work with our colleagues at Gene2Phenotype, PanelApp Australia, the Gene Curation Coalition and disease experts to share gene-disease curations throughout the international rare disease and cancer communities.
Artificial Intelligence (AI) has great potential for improving the efficiency of our gene-disease evidence gathering and curation processes and we are exploring its use. For example, we have conducted a trial using Machine Learning to find publications relating to the ‘Intellectual disability’ panel. Using these tools, we were able to add twice as many new green gene-disease associations to the panel over 6 months compared to crowd-sourcing updates alone as explained in our Accelerating biocuration webinar.
The PanelApp team at Genomics England are excited to continue to build on the successes of the last 10 years and to help shape the genome analyses of the future.
Want to read more about the work happening at Genomics England? Check out our other bioinformatics blogs.


