Developing an algorithm to de-identify pathology reports: part 2
By Andreia Rogerio and Stanley Ng onIn Part 2 of this blog series, Andreia Rogerio and Stanley Ng, Applied Machine Learning Engineers at Genomics England, explore the algorithms used in the de-identification pipeline, explaining how and why each of them was chosen.
The initial segment of this two-part series provides an overview of the structure of the de-identification pipeline. In this second instalment, we will delve into the specifics of the pattern-matching and NLP algorithms used in the pipeline, offering insights into the selection process and the rationale behind each choice. We will also share a link to an interactive demo of our de-identification tool.
The big picture: a two-stage process
The pipeline is designed to process and redact sensitive information from pathology reports presented in simple text format (Figure 1). This process is important for researchers to access de-identified pathology reports, which contain information not available elsewhere. At Genomics England, we support access to our data through the Trusted Research Environment (TRE).
The first step of the de-identification pipeline is a detection stage. This stage is composed of several elements: a Microsoft Presidio model1, regex modules, fine-tuned Natural Language Processing (NLP) models for the Name Entity Recognition (NER) task, and a pre-defined list of words that do not require redaction. Additionally, a list of patient metadata is used to ensure that all known patient identifiable data is removed.
Following the detection stage, the data proceeds to a redaction stage. Here, the pipeline identifies the words marked for redaction from the previous stage, and substitutes each occurrence with appropriate redaction characters.
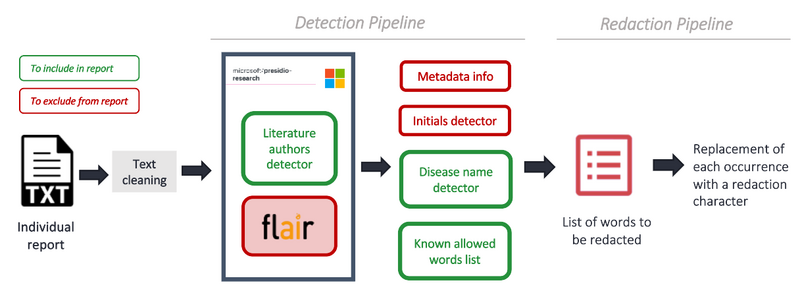
Figure 1: De-identification pipeline, consisting of two main stages: detection and redaction.
Built from the ground up
The pipeline was built iteratively starting with a simple approach - the off-the-shelf Microsoft Presidio model represented by the grey box in Figure 1. We gradually incorporated more complex and tailored modules as we observed which entities were being detected and which ones were being missed.
This iterative process was guided by manual validation with Ground Truth2, an AWS service for data labelling (explained in part 1 of this series), ensuring the accuracy of our pipeline.
1. Patient identifiable information
One of the key modules in our pipeline is the metadata filtering module. This module uses existing metadata about the reports to confidently redact patient names, NHS numbers, dates of birth, and patients’ addresses and phone numbers.
To guarantee complete removal of personally identifiable information, we only apply the de-identification pipeline to reports that meet our eligibility criteria. In this context, eligible reports are those that come with complete high-quality metadata. These allow us to confidently assert that an exhaustive search filter (commonly called brute-force approach) will redact all sensitive information about the patient.
Due to the lack of completeness of the addresses we can only redact approximately 60% of the reports, which resulted in the release of 46,556 reports out of the approximately 80,000 available.
Using the metadata for an exhaustive search is not straightforward, in many cases, the data might be represented in different ways. For example, the same date or address might be written in different formats, so custom RegEx expressions are needed to ensure that we redact all personal data while keeping important information visible, such as diagnosis or treatment dates.
2. Staff names and initials
The detection of staff’s names and initials in our pipeline presents a unique challenge. Unlike patient identifiable data, we don't have metadata for staff names and initials, which necessitates the use of machine learning techniques.
To tackle this, we used Microsoft Presidio and integrated several pre-trained NLP models in this software. After comparing their performances, Flair3 emerged as the most effective at detecting personal names, achieving recall values of 95.8% for staff names and 87.1% for staff initials in a dataset of 400 randomly selected reports for manual inspection.
Flair was first introduced in 2018 as a monolingual (English) model based on a bi-LSTM (bi-directional long short-term memory network) architecture, pre-trained on Wikipedia data. Since then it has seen several improvements, and FlairNLP is now a framework for experimentation with language models, that includes several versions of models for NER tasks.
The most recent SoTA NER model in the FlairNLP framework is FLERT XLM-R4 (2020); it contains a CRF (Conditional Random Field) layer, which allows the model to consider the predicted labels for the neighbouring words when making a particular prediction, which is helpful in NER tasks (e.g., detecting a full name or an address). The character-based embedding approach used by this model (i.e., the method it uses to convert each word into a numerical representation) is particularly good at modelling subwords, suffixes, and prefixes - crucial elements for an NER task.
Currently, other cutting-edge models are available that surpass FLERT in performance. In particular, ACE + document-context and BERT-MRC+ DSC are the current SoTA models for the ConLL20035 and Ontonotes v5 (English)6 datasets respectively.
At the time of writing, FLERT ranks 2nd in the ConLL2003 dataset, though it wasn't assessed using the Ontonotes v5 dataset. Nonetheless, for the purposes of this de-identification project, the Flair framework has proven to meet our desired standards in terms of machine learning performance (i.e., the required recall for each entity) at a good speed, making it a worthwhile choice.
2.1. Machine learning names detector performance compared to Amazon Comprehend:
To validate the effectiveness of our machine learning solution, we conducted a comparative analysis with AWS Comprehend7. This is a well-known NLP service that uses machine learning to analyse and extract information from text. We used a dataset of manually labelled cohorts, specifically focusing on Acute Myeloid Leukemia (AML) and Testicular cancer types from the National Cancer Registration and Analysis Service (NCRAS)8.
The results of this comparison were quite revealing. Our detector not only overlapped with AWS Comprehend in detecting most entities, but it also identified additional entities that AWS Comprehend missed. Specifically, our model found 304 entities in the AML cohort, and 146 in the Testicular cancer cohort that were not detected by AWS Comprehend. Upon closer examination, many of the names found by Flair and not by AWS Comprehend were initials that easily resemble organisation’s acronyms (e.g., ‘WFH’ or ‘JS’) and are a known challenging use-case in name entity recognition tasks.
Conversely, AWS Comprehend only found 4 entities in the AML cohort that were not found by our detector.
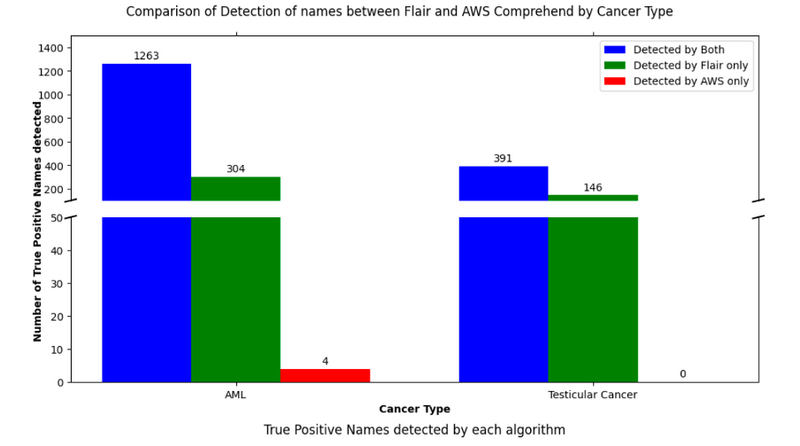
Figure 2: Comparative performance of Flair integrated in Presidio vs. Amazon AWS Comprehend. The prediction task is the detection of names in 2 independent cohorts (AML and Testicular cancer). Note that for the y-axis 1) the values between 50 and 150 are omitted; and 2) the scales of each subplot (top and bottom) are different, for visibility purposes.
3. Redaction
The final step in our pipeline is the redaction process. This is where we take the sensitive information that has been identified and mask it to protect patient privacy. We accomplish this by replacing each identified sensitive character in the text with ‘¤’ to differentiate from other characters while keeping readability.
The process is carried out by a function that operates on the principle of string replacement. It loops over each sequence that has been flagged for redaction and replaces it with a string of the redaction character that is of the same length as the original sequence. This maintains the structure and length of the text while obscuring the sensitive data, enabling compliant data use and analysis in our Research Environment.
Key learnings
1. Human Validation is Key: Throughout the development and validation of our pipeline, one of the most important lessons we've learned is the irreplaceable value of human engagement. While automated processes and machine learning models can be incredibly powerful, they are not infallible.
2. Medical Data is Highly Heterogeneous: Another key insight is the complexity associated with redacting personal identifiable information from medical free-text documents. In these documents, patient’s personal information is not organised in a structured format like headers (commonly located at the top of medical reports). This means we can only ensure it is removed by processing and analysing the full length of each report. To de-identify unstructured data, the best approach complements manual inspection with deep learning techniques.
3. Regex efficiency can surpass ML: We've discovered that machine learning may not always provide the specificity needed for some cases, in which simple regex patterns can be employed. When no metadata is available (e.g., for clinical staff) and the required redaction level is below 100%, machine learning provides the best automated solution. However, if 100% redaction is required, an exhaustive search approach (using each patient’s metadata) is the only automated method able to achieve it.
Now
that we have covered the intricate workings under the hood, it's time to
see how well it does. We prepared a demo to mimic how the real pipeline works, and made it available through HuggingFace Spaces. We invite you to give it
a try and witness its capabilities in action, with your own data.
Next steps
One of our immediate next steps is to leverage the Postcode Address File (PAF) database from Royal Mail. This comprehensive address database will allow us to redact addresses more effectively through an exhaustive search approach.
Additionally, we plan to implement an automated regular validation process. This process will involve assigned staff members periodically reviewing a representative sample of the dataset, including new data when available. They will manually label this representative sample, providing a ground truth against which the performance of our models can be continually measured.
Acknowledgements
Firstly, our acknowledgement goes to the participants of the 100,000 Genomes Project, whose data has been at the core of this project. Special mentions to Francisco Azuaje for steering the technical aspects of the ML work stream, and to Prabhu Arumugam for his insights into Pathology data interpretation and best practices.
We also thank the CCDT (Cancer Clinical Data Team) team members, in particular Georgia Chan, James Micklewright, Marko Cubric, for the time and effort dedicated to labelling reports. And reiterating our previous acknowledgements, our sincere thanks to the Data Protection and Ethics teams for their invaluable guidance and support.
References
- Microsoft Presidio: https://microsoft.github.io/presidio/
- Amazon SageMaker Ground Truth https://aws.amazon.com/sagemaker/data-labeling/
- FLERT publication: https://arxiv.org/pdf/2011.06993v2.pdf
- Flair github repository: https://github.com/flairNLP/flair
- CoNLL-2003*: W03-0419.pdf (aclanthology.org)
- OntoNotes v5*: OntoNotes Release 5.0 - Linguistic Data Consortium (upenn.edu)
- AWS Comprehend: https://aws.amazon.com/comprehend/https://aws.amazon.com/comprehend/
- NCRAS: https://www.gov.uk/guidance/national-cancer-registration-and-analysis-service-ncrashttps://www.gov.uk/guidance/national-cancer-registration-and-analysis-service-ncras
* CoNLL-2003 and OntoNotes v5 are datasets commonly used as a reference standard for NER tasks as they contain thousands of public documents (like news articles and blogs) labelled by the Linguistic Data Consortium (LDC)


