Extracting bladder tumour characteristics from pathology reports using Large Language Models
By Marko Cubric, Alastair Hall, Samuel Barnett, Charlotte Jennings, Jon Griffin and Cong Chen onAt Genomics England, we hold data for over 15,000 participants with cancer in the National Genomic Research Library (NGRL). Whole genome sequencing for these participants can lead to improvements in clinical treatment and outcomes, and we already know of various genetic markers that tell us about the source of a cancer or what treatments might be appropriate. For researchers to make sense of the data in the NGRL, they need to combine it with clinical data, such as judgements from clinicians, so that they can validate where genomics adds value.
To do this, we have access to over 70,000 clinical pathology reports for cancer participants. These reports are potential gold mines for researchers as they contain valuable information for prognosis that is not always present in available secondary data. This blog explores the benefits of applying Large Language Models to extract information from pathology reports, which allows us to quickly curate bespoke structured data from unstructured reports based on specific researcher needs.
Investigating bladder cancer
Through discussion with our internal clinical pathologist, we identified a user in the Bladder Cancer Research Network looking to stratify cancer patients into different pathological groups according to various tumour characteristics.
In the case of bladder cancer, there are groups that have different prognosis, treatment options and general clinical trajectories. These groups are largely defined by the stage and grade of their tumour and the presence or absence of Carcinoma in Situ (CIS). Being able to stratify bladder cancer participants into these categories would enable the research user to perform analysis on genomic changes and understand how these changes are linked to their clinical data.
When the user began working with bladder cancer data in 2017, there was not enough secondary clinical data available to perform their research combining both clinical and genomic data. Despite the sparsity of this data, a lot of the relevant information for research could still be found in patients’ pathology reports. However, as these are primary health records containing personal identifiers, Genomics England does not release these to researchers.
Only when our first pathology report de-identification algorithm was developed in 2023 was our user able to curate a dataset suitable for their research needs. Although an improvement, this was a time-intensive task that involved the user having to read through over 400 de-identified pathology reports and manually extract relevant tumour characteristics from the text.
In addition, due to data protection constraints, not all available pathology reports met the required governance standards to be released into our Research Environment. This resulted in a smaller set of reports that our user could use, and thus a less complete dataset for them to work with. Figure 1 shows a typical bladder cancer pathology report with relevant tumour characteristics highlighted.
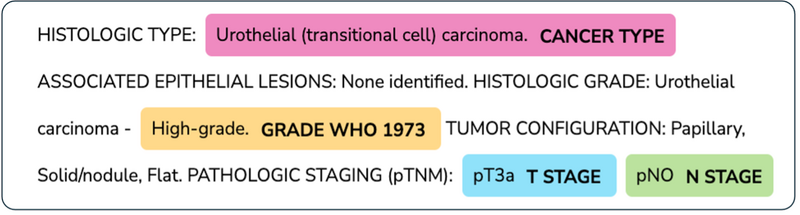
Figure 1: Example TCGA pathology report with LLM extracted bladder tumour characteristics highlighted
For this use-case, the complete set of relevant tumour characteristics to extract from the reports are described in following table. All these characteristics help researchers to stratify patients for their research.
|
Characteristic |
Description |
|
Grade (WHO 1973) |
The level of aggressiveness the cells demonstrate, ranging from Grade 1 (lowest grade, the cancer cells look like normal cells) to Grade 3 (highest grade, the cancer cells look very abnormal). |
|
Grade (WHO 2004/ISUP) |
‘Low grade’ bladder cancer means that your cancer is less likely to grow, spread and come back after treatment. ‘High grade’ means your cancer is more likely to grow spread and come back after treatment. |
|
T Stage |
T stage refers to the size and extent of the main tumour. The main tumour is usually called the primary tumour. |
|
N Stage |
The N refers to the number of nearby lymph nodes that have cancer. |
|
Carcinoma In Situ (CIS) |
A condition in which abnormal cells that look like cancer cells under a microscope are found only in the place where they first formed and haven't spread to nearby tissue. |
|
Cancer type |
The cancer type of the primary tumour sample |
|
Cancer subtype |
The cancer subtype of the tumour sample based on the appearance of the tumour under a microscope |
What was our approach?
The characteristics defined in the above table can also be considered as “entities” in a Natural Language Processing (NLP) context. NLP is the field of machine learning that deals with human written text. The task of extracting these entities from free-text is commonly known as Named Entity Recognition (NER). See the Turing comprehensive guide to named entity recognition for further explanation.
Traditional methods such as off-the-shelf NLP tools like AWS Comprehend, regex-based approaches, and standard NLP packages like SpaCy can fall short in NER tasks without substantial customisation. They can require extensive tuning to perform adequately, making them labour-intensive to adapt to the requirements of each specific use case.
Large Language Models (LLMs) are a more recent generative AI approach that can also be harnessed to solve classical NLP tasks. They are a type of neural network based on the transformer model, and are trained on vast amounts of data. In the context of NER, they prove to be a viable alternative to traditional methods due to several key advantages:
- Generalisability: LLMs can adapt to different contexts, reducing the need for labour-intensive customisation and allowing more efficient deployment across diverse report types.
- Efficiency in Development: Pre-trained architectures enabled a rapid transition from development to implementation, cutting down on fine-tuning time.
- High Initial Performance: Models like Claude 3 Sonnet showed strong out-of-the-box performance, accurately interpreting clinical language with minimal adjustments.
This work builds on an initial proof of concept which tested LLMs including Mixtral 7xb, Llama2 and Claude both self-hosted and serverless, alongside AWS Bedrock-hosted models. Claude on Bedrock emerged as the top performer, thanks to its succinct responses and fast processing times.
In this project, we tested Claude 3 Sonnet on a new set of 100 pathology reports labelled by pathologists with the tumour characteristics of interest for validation. We used 3 rounds of collaborative prompt engineering to improve the quality of the LLM extraction. This structured approach effectively harnessed Claude’s adaptability, achieving high precision with minimal manual intervention and marking a significant improvement over traditional NLP techniques for both development time and performance.
How did we assess performance?
In our case, each entity we were extracting fit into a categorical group of possible values. This allowed us to compute performance metrics using a multiclass approach for each entity.
When reviewing our labelled data, we identified that many of the class sets were moderately or significantly imbalanced. To account for this, we used a combination of standard performance metrics: precision and recall. We calculated these metrics at the class level, taking a weighted average across classes for a single entity to obtain a more balanced a meaningful assessment. We also used confusion matrices to identify any classes that were underperforming relative to the class set. A brief description of the performance metrics used are outlined below.
- Precision measures the proportion of correctly extracted entities out of all entities identified.
- Recall measures the proportion of relevant entities correctly identified from the total number of relevant entities.
We avoided other standard metrics like accuracy, because it can be misleading in imbalanced datasets, often skewed by the majority class. While F1-score combines precision and recall into a single measure, it obscures which of the two metrics is dominating the overall performance.
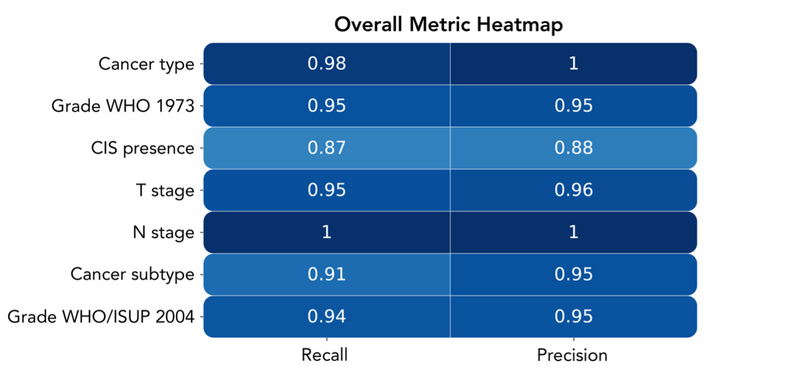
Figure 2: Heatmap of overall performance by entity
Figure 2 demonstrates high performance across all metrics for all entities, showing that the extracted information is reliable for the researcher’s use-case. Some of the entities we were extracting contained highly imbalanced classes, most notably cancer type, N stage and cancer subtype. This led us to use weighted average metrics across all entities to avoid over-representation of low prevalence classes in our aggregate metrics.
Discussion
This project successfully demonstrated the power of using LLMs in extracting entities from pathology reports. The initial aim of the project was to achieve high quality and correct extractions from pathology reports which is demonstrated in Figure 2.
In working through this use-case, we have also developed a generalisable application that easily harnesses LLMs to extract requested information from pathology reports. With this tooling, it is far easier to track prompts, results, and cohorts of reports in a scalable way to empower us in future use-cases.
An interesting and consistent trend we observed was related to way the LLM interpreted the provided labels presented in the prompt. We noticed that subtle changes could significantly impact the overall performance.
For example, when developing the part of the prompt that handled cancer grade from the 1973 classification, the model performed worse when asked to extract grades ‘1’,2’ and ‘3’ versus grades ‘G1’, ‘G2’, ‘G3’ despite having the same meaning. We think this is due to the ambiguity of asking the LLM to look for integers as opposed specific values with intrinsic meaning. By asking the model to look for integers, we were also asking it to make an association to a concept, which may have added complexity to the task it must perform, reducing the quality of its response.
We also faced a similar issue when trying to extract CIS status. This entity is reported in the text as ‘Present’ and ‘Not Present’. We think this caused the model some confusion where it conflated whether CIS was ‘Present’ or ‘Not Present’ in the tumour, or whether CIS was present or not present in the text.
Now that we have successfully extracted data for our user, we are focusing on applying this approach to different use-cases. We are currently working on a project to extract HER-2 statuses across the whole cohort of clinical cancer pathology reports. We hope that this use-case will demonstrate the generalisability and scalability of our approach.
Acknowledgements
Thank you to all the Genomics England colleagues who have helped us with this project. In particular thanks to Andreia Rogerio, Georgia Chan and the wider Multimodal squad for their guidance and expertise.
We would also like to thank colleagues at Amazon Web Services: Prabhu Arumugam, Matt Howard, Lou Warnett, Cemre Zor, Michael Mueller and Anastasia Tzeveleka for their help in the initial stages of development.


