Renewing our knowledge of gene-disease associations for rare conditions
By Eleanor Williams onSenior scientific curator, Eleanor Williams, explains the importance of keeping gene-disease association panels up to date, and how we collect evidence to do this.
Whole genome sequencing (WGS) is increasingly used for the diagnosis of human genetic conditions. However, only a minority of the millions of gene variants identified by WGS are likely to cause the affected person’s condition.
In a previous blog, our bioinformatician Kevin Savage describes the automated variant triaging framework called Tiering. This framework is used to create a short list of prioritised candidate variants from WGS for clinical laboratories to assess.
After a variant has passed the initial filtering steps (being rare, resulting in protein alteration, and following expected segregation within the patient’s family), the Tiering process screens virtual lists of genes, known as panels.
This screening reveals whether the variant is in a gene that has previously been associated with a similar condition to the one being investigated. If there is a match, and if the inheritance pattern in the patient's family is the same as other reported cases, then the variant is considered for in-depth interpretation.
The Tiering blog post describes a retrospective analysis of variants from the 100,000 Genomes Project that were not originally prioritised but turned out to be diagnostic. The team determined that the primary cause of a variant not being prioritised initially was ‘insufficient evidence’ for gene-disease association at the time of the analysis.
This shows that keeping gene panels up-to-date is a vital aspect of the work that Genomics England does, directly impacting the diagnostic rates for genetic conditions.
PanelApp knowledgebase of gene-disease associations
At Genomics England, information about gene-disease associations is stored in a public knowledgebase called PanelApp.
Phenotypically related gene-disease associations are grouped together into virtual panels e.g., the skeletal dysplasia panel, or the intellectual disability panel. Specific panels are then applied as part of the variant Tiering pipeline, based on the clinical features of an affected individual.
Scientific curators review any evidence that indicates a gene might be associated with a particular condition. We use a so-called 'traffic light' system to designate the strength of the evidence. The broad-term colour ratings are shown in Figure 1.

Figure 1: PanelApp traffic-light system for evidence level
Only genes with a 'green' rating are used in the Tiering process; the amber and red rated genes are on the panel to show that the evidence has been looked at, but is not currently strong enough to regard the gene as diagnostically reportable. A more detailed description of the criteria for rating a gene green is given here.
Panels may also contain information about haploinsufficiency or triplosensitivity regions (copy number variants) and short tandem repeats associated with disorders.
Panels vary in size; some have only a few genes e.g., the panel for Brugada syndrome shown in Figure 2, and others are much larger, such as the Early onset or syndromic epilepsy panel with over 800 genes. Panels are versioned so that the content of a panel at the point of analysis can always be retrieved.
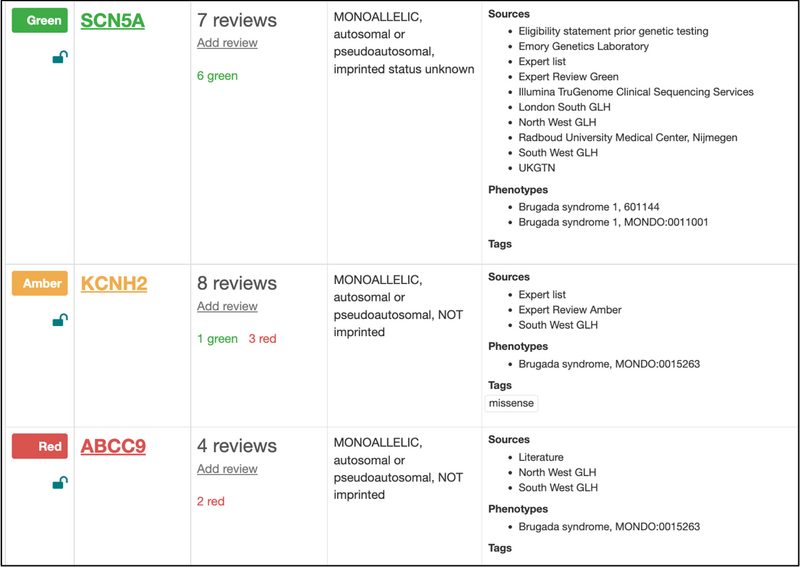
Figure 2: Partial view of the genes on the Brugada syndrome panel, showing gene symbols and ratings, modes of inheritance and phenotypes.
Panels used in the 100,000 Genomes Project and the NHS Genomic Medicine Service
Around 175 different gene panels were used in the WGS analysis of patients with rare conditions in the 100,000 Genomes Project.
Panels were initially created by searching high-quality gene–disease information sources. Then, experts in the disease field, from both the UK and other countries, were invited to review the panels. Further information on this process can be found in Rueda Martin et al. 2019, Nature Genetics, 51:1560–1565.
The NHS Genomic Medicine Service (GMS) was launched in 2018 and WGS was chosen as the genomic analysis method for several clinical indications. This was building upon the success of the 100,000 Genomes Project. Panels were developed in PanelApp for this service, and all panels were reviewed by disease experts from within the NHS.
In addition to WGS panels, PanelApp holds panels of genes that relate to other genomic tests listed in the National Genomic Test Directory, creating a single searchable resource.
PanelApp shows the current versions of NHS GMS panels as well as other panels from the 100,000 Genomes Project. To see and search for just the NHS GMS panels, or for specific versions approved for use in genomic analysis by the NHS, visit the GMS Panels website.
How are panels kept up-to-date?
It is the job of the curation team at Genomics England to gather and assess new evidence for gene-disease associations.
A unique feature of the PanelApp webtool is that experts in rare conditions can add and review gene-disease evidence. This crowdsourcing of knowledge has proven invaluable in keeping the gene panels up-to-date, and is used by many NHS clinicians and clinical scientists to propose changes to the GMS panels.
PanelApp has over 300 registered expert reviewers from around the world who have provided reviews. Throughout 2022, we received 394 reviews, of which 51% were from NHS clinicians and scientists.
We evaluate the evidence provided in each review, and may also add the gene to additional panels depending on the phenotype associated with the reported condition.
The curation stage can be quick in cases with clear reporting of patient phenotypes, analysis methods, segregation data and genotype information. However, it may take longer if for example evidence is coming from several publications and/or the phenotypes are variable or complex.
In addition, we gather information from sources such as key scientific journals, other gene-disease association databases (e.g. OMIM, ClinGen and GenCC), and compare panels from genomic services like PanelApp Australia. Feedback of diagnostic variants identified in the 100,000 Genomes Project and the GMS is also becoming an important source of evidence for gene-disease associations.
Final review and signoff of panel content to be adopted into NHS service is done through the NHS Genomic Laboratory Hubs and their associated colleagues. This is under the governance of the national genomic test directory evaluation process.
The first step is to compile a report of proposed changes to the panels. Evidence is collated in PanelApp, either from reviewers directly, by curators adding information, or from proposals for new clinical indications. It can also be gathered from amendments to existing clinical indications submitted directly to the Genomics Unit at NHSE. Genomics England curators then send reports of proposed updates to the NHSE Genomics Unit.
Once evaluation of the proposed changes is complete, the panels are updated accordingly, and new versions of panels are approved for use in the rare disease analysis pipeline and in NHSE Genomic Laboratory Hubs.
Since the first versions of GMS panels were signed-off, we have added a further 1,936 green entities on the latest agreed panels. We have also improved the accuracy of existing data by demoting 196 genes that did not meet the criteria for a diagnostic-grade green rating upon reassessment, and we have updated the mode of inheritances of 232 green genes. We have also added 15 new NHS GMS panels in this time.
The new approved versions are displayed in the GMS Panels website. The process is shown in Figure 3 below.
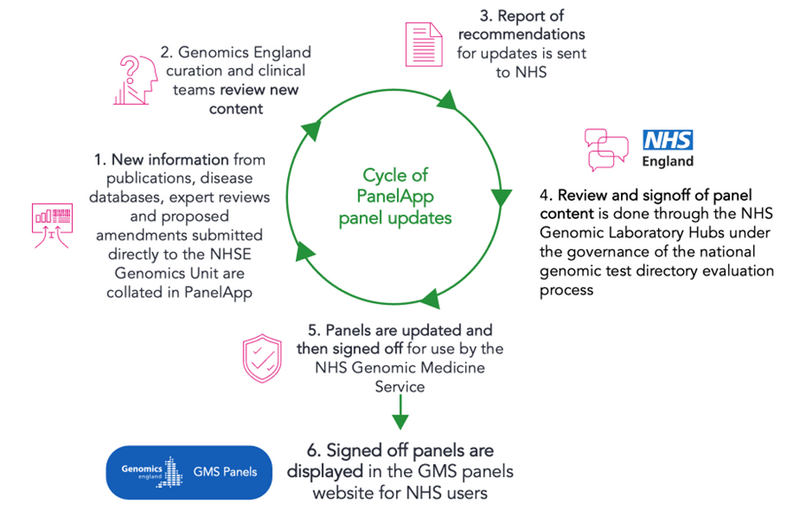
Figure 3: Cycle of updates of gene panels used in the NHS Genomic Medicine Service and subsequent display in the GMS panels website.
The success of automated variant prioritisation approaches is largely dependent on the accuracy and comprehensiveness of gene-disease association datasets.
The PanelApp knowledgebase provides a means of collating this information. It acts as a central point of reference for gene-disease evidence for all NHS genetic laboratories across England, and serves as a public source of information for clinical and research laboratories world-wide.


