Increasing gene coverage for developmental disorders in PanelApp using the Gene2Phenotype database
By Achchuthan Shanmugasundram onIn this blog, Achchuthan Shanmugasundram, Scientific Curator at Genomics England, describes using a gene panel from an external resource to improve gene coverage when testing patients in the NHS Genomic Medicine Service (GMS) in England.
Gene panels are virtual lists of gene-disease associations that are grouped into specific disease areas. They are used to filter and prioritise the millions of gene variants that are identified as different to the reference genome in patients with rare genetic conditions. Through this prioritisation, the number of variants can be reduced to a handful, which can then be manually assessed by clinical scientists.
There are ever more gene-disease associations being reported in the scientific literature. This evolving knowledge should be used to improve the diagnostic yield in patients tested using whole genome sequencing analysis. There are several resources that capture these associations, and sharing of knowledge between these resources will benefit affected patients across the globe.
Genomics England PanelApp and GMS Panels
Whole genome sequencing performed by Genomics England as part of the NHS GMS uses virtual gene panels hosted in the Genomics England PanelApp. In a previous bioinformatics blog, we discussed the process of curating gene-disease associations within virtual panels, and how panels are kept up to date in collaboration with the NHS.
To summarise, gene-disease associations are rated using an evidence-based traffic light system from red, through amber, to green, with green indicating a diagnostic level of evidence. All changes to the diagnostic grade gene content of panels are agreed with the NHS before being implemented in PanelApp. New versions of panels are ‘signed-off’ for use, and are displayed in a PanelApp companion site, the NHS GMS Signed Off Panels Resource.
Developmental Disorders (DDG2P) panel and its use in the GMS
The panels hosted in PanelApp and used in the GMS are defined by the NHS England National Genomic Test Directory and are curated by the PanelApp team. The only exception to this is the Developmental Disorders (DD) panel, which is obtained from the Gene2Phenotype (G2P) database (named ‘DDG2P’ in PanelApp) and has been integrated without further curation due to the large size of the panel.
Although the DDG2P panel partially overlaps with other GMS panels, it has much broader scope than any existing panels, and therefore provides some unique gene-disease assertions.
Like PanelApp, the G2P database from the European Bioinformatics Institute hosts virtual panels for various disease areas. The DD panel from the G2P database is used by NHS Scotland for testing patients diagnosed with developmental disorders, thereby serving as a complementary resource to PanelApp.
The scope of the DD panel is defined as genes associated with ‘severe undiagnosed neurodevelopmental disorder and/or congenital anomalies, abnormal growth parameters, dysmorphic features, and unusual behavioural phenotypes’ (Wright et al. 2015, Lancet).
In the GMS, the DDG2P panel is a component panel of the ‘R27 Paediatric disorders’ superpanel. This is the most requested test within the Rare Disease GMS, having been applied in over 8,000 cases since the inception of the NHS GMS.
The importance of updating the panel
Since the previous DDG2P PanelApp update in November 2019, G2P has changed the terminology used for gene-disease ratings (‘confidence category’ in G2P), modes of inheritance (MOIs) and modes of pathogenicity (MOPs) to align with the Gene Curation Coalition (GenCC) nomenclature.
The GenCC coalition aims to harmonise gene-level resources and has developed standardised terminology to curate gene-disease associations. Further details of this body can be found in DiStefano et al. 2022, Genetics in Medicine. However, though Genomics England PanelApp is a member of the GenCC, we have not adopted their nomenclature due to constraints within our analysis pipelines.
G2P previously had 3 different gene-disease confidence ratings (possible, probable and confirmed) which we mapped into PanelApp to red, amber and green respectively. However, G2P now has four rating categories aligning to GenCC nomenclature.
Figure 1 shows how the previous G2P ratings were mapped to PanelApp ratings, and how G2P has mapped them to the new GenCC ratings. The new ‘moderate’ category that lies between ‘limited’ and ‘strong’ does not have a direct mapping from the previous G2P confidence categories. The ratings of ‘moderate’, ‘strong’ and ‘definitive’ are used for prioritising variants from patient genomes by NHS Scotland, as genes with these ratings are considered to have diagnostic potential.
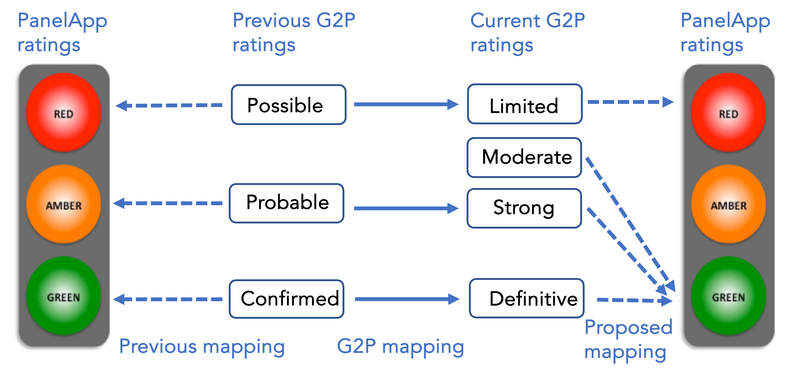
Figure 1: The previous G2P evidence ratings mapped to the PanelApp traffic light system of ratings and to the current G2P ratings. It also shows the future mapping between current G2P and PanelApp ratings.
The process of mapping ratings between the two resources
The GenCC/current G2P ratings are defined based on the number of published manuscripts reporting the gene-disease association. PanelApp ratings, however, are based on the number of unrelated cases and functional evidence from experimental studies.
Although the guidance for ‘moderate’ rating is that the body of evidence is not large and may be based on one key paper, this paper may report 3 independent cases with plausible disease-causing variants in the gene, therefore fulfilling our criteria for green rating. The mapping of ratings between these resources is therefore not a straightforward process.
The DDG2P panel is one of the largest panels used in the GMS with over 1,900 gene-disease associations. Re-curating all disease associations between the 2 rating systems would be a labour-intensive process and therefore is not feasible. Instead, we want to update this panel regularly in a mainly automated manner, while still maintaining the accuracy of the confidence levels.
2 of these ratings were straightforward to map. The ‘definitive’ rating is quite stringent, with the requirement of repeated demonstration of the gene-disease association in both the research and clinical diagnostic settings, hence we can confidently map this to PanelApp’s green rating. The ‘limited’ rating is equivalent to red. We then wanted to conduct an evidence-based assessment of how ‘strong’ and ‘moderate’ ratings should be mapped to PanelApp ratings.
To do this, we manually curated a subset of genes from DDG2P, with the 2 ratings based on the DD panel scope. All genes with ‘strong’ rating and 90% of sampled genes with ‘moderate’ rating could be rated green (there were under 40 genes in the ‘moderate’ category when this work was done). This analysis shows that our previous mapping of G2P ‘probable’ to PanelApp ‘amber’ was likely too conservative.
After consultation with the G2P team and as part of the National Genomic Test Directory evaluation process, we concluded that both ‘strong’ and ‘moderate’ should be mapped to green (Figure 1) so that the data could be imported in an automated way in the future without manual curation of each individual gene-disease association.
The impact of update on the GMS
Following the decision on how to handle the conversion of gene-disease ratings between the two resources, we updated our DDG2P panel using the July 2023 static release of the DD panel from G2P. This version of DD panel had 2,646 gene-disease associations, covering 2,336 genes (some genes have more than one disease association). Of these, associations for 423 genes had been added since the last update of the DDG2P panel. After mapping to PanelApp ratings, there are 2,014 green genes, which is an increase of 820 green genes from the previous panel version.
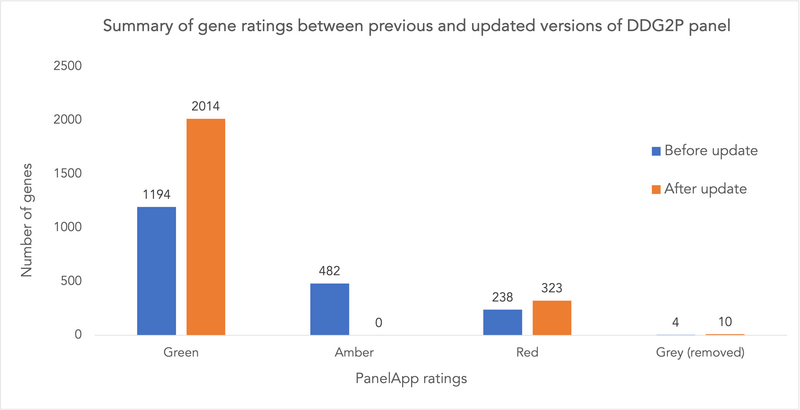
Figure 2: Summary of gene ratings between the previous and the updated versions of DDG2P panel in PanelApp.
Reflecting other changes in G2P, we updated MOIs for 60 genes and demoted 8 genes to grey as they lacked any evidence for the disease association and have now been removed from G2P.
Overall, the updated DDG2P panel adds 250 additional green genes to the Paediatric disorders superpanel. 95 of these genes are not green on any GMS panels, for reasons such as some of them not fitting into the scope of any of the other GMS panels.
Following this update, we have now released a new signed-off version of the DDG2P panel (version 4.0) for use in the GMS in our May 2024 data release.
Through this work we now have a protocol, in agreement with G2P and NHS England, for reimporting the DDG2P panel which can be used to perform annual updates.
It highlights the challenges of interoperability between resources using different rating systems and terminology, as well as the benefits of publicly shared data. This update will potentially lead to an increased diagnostic yield for patients tested for paediatric disorders through the GMS.
We would like to thank our colleagues in G2P and NHS England for their help with this work.
And finally...
Save the date! Register for the annual Genomics England Research Summit on 9 July 2024.
To learn more about the work happening at Genomics England, check out our other bioinformatics blogs.


