Launching a global standard resource for cancer research
By Prabhu Arumugam and Nour Merzouki on
It is increasingly accepted that to better diagnose and predict cancer, we need to understand it at both a molecular (genomic) and a spatial (histological and radiological) level. That’s why, as part of our Cancer 2.0 initiative, we’re building the world’s largest whole genome sequence (WGS) multimodal cancer research platform - combining genomic, clinical, and imaging data.
The first of its kind, this platform builds upon the success and productivity of previous datasets, such as the Cancer Genome Atlas (TCGA). It aims to make the most of the community we have at Genomics England, along with our population, clinical, and research work.
The platform brings together the latest generation of genomic, longitudinal, clinical, and imaging data at an unprecedented scale. It resides in a secure cloud-based research environment, with scalable compute infrastructure with a range bioinformatics and AI/ML analytical tools, and the ability for researchers to bring their own.
Our aim is to enable academic and industry scientists from around the world to use this data to transform our understanding of tumour biology, allowing the development of novel targets and treatments, and improvements in diagnosis, prognosis, and treatment selection for patients.
Progress on the platform is advancing rapidly. The pathology component is already 3 times the size of TCGA, and has been scanned at twice the resolution. It is growing fast as we import digitised imaging data.
We already have some early adopter clinical and biopharma users exploring the data, and plan to launch it with the wider research community early next year.
A big need and a unique opportunity
In cancer, cross-talk between the molecular and morphological features of a tumour, the surrounding micro-environment, and the immune system, can impact on diagnosis, prognosis and response to specific therapies.
For example, we know that PD1 expression on the tumour cell surface, along with the presence of lymphocytes in the tumour micro-environment, are both required for a successful response to immune checkpoint inhibitors. Combining molecular and spatial insights such as these have tremendous research value, potentially identifying new druggable protein targets or new mechanisms of action. They can also help to better stratify patients for clinical trial inclusion.
Current research insights are generated by existing, separate technologies, such as sequencing and digitisation of histology slides. However, there is increasing interest in combining digital pathology and radiology findings with molecular signatures. This type of multimodal data will unlock new areas of genomic research, and could open doors to genetic mutations being identified from patient imaging.
Our new cancer research platform will introduce multi-modality for research, by combining genomic, clinical, and imaging data. The digital pathology and radiology images will form an image library to sit alongside the genomic and clinical data for each participant within the Genomics England Research Environment. This platform will be the first of its kind, accelerating cancer and genomic research by providing patient images which are digitised, de-identified, annotated, and prepared for users to conduct deeper analysis.
To realise this landmark project, we have been joined by key partners with leading expertise in the new modalities of data and analytics that are driving the transformation of cancer research. Amongst these partners are the National Pathology Imaging Co-operative (NPIC) in Leeds, who are digitising hundreds of thousands of pathology slides from across more than 20 NHS trusts; Insitro, who are providing a semantic structure for the imaging data to enable its analysis with machine learning; and AWS, who are collaborating on the cloud infrastructure to make both the data and tools available to users.
Through a series of blogs, we will share our progress in building the platform, and introduce you to some of the people making it happen and putting it to use. First up, Nour Merzouki, Programme Manager at Genomics England, shares more about our image retrieval process.
How we’re collecting imaging data
As part of the image retrieval process, we are collecting pathology and radiology imaging data, and combining it with clinical and genomic datasets. We are collecting the imaging data from NHS sites; for pathology we are collecting glass slides from 100,000 Genomes Project participants, and scanning them at NPIC.
NPIC is a Centre of Excellence in digital pathology and AI, focused on the clinical deployment of digital pathology in the NHS. Over the next two years, working with NPIC, we will generate whole slide images for the entire 100,000 Genomes Project cancer cohort, linked to curated pathology data.
This cohort includes more than 15,000 participants recruited from 84 hospitals across England. It includes people with tumours from 20 solid cancer subspecialty areas, all of whom were prospectively recruited, sampled, and sequenced with the same protocols.
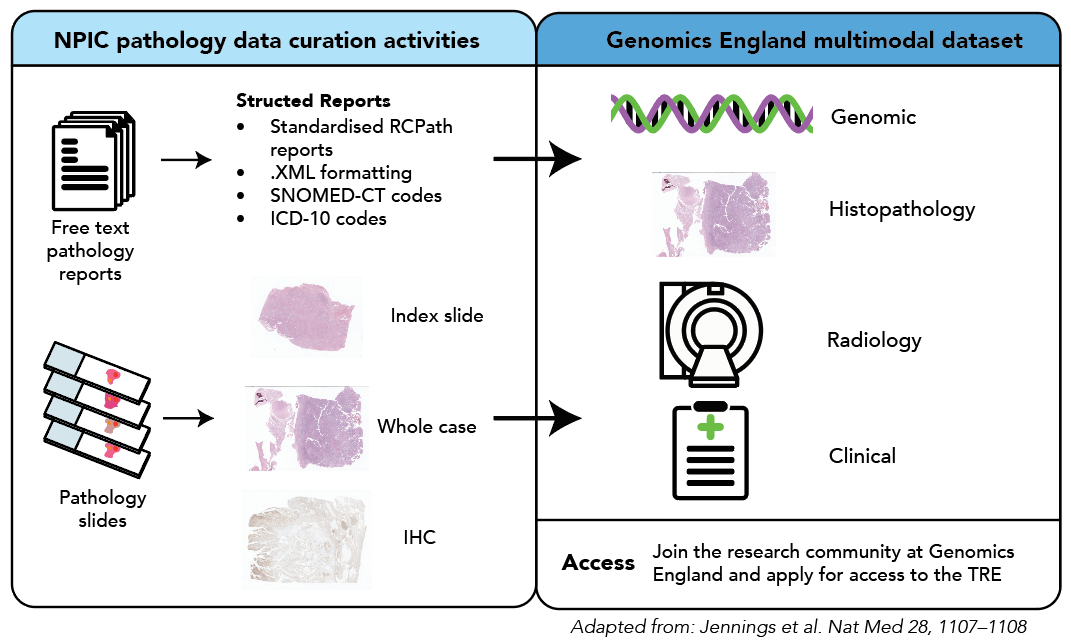
To maximise the utility of the data for biological discovery work, where possible, the whole pathology case for each tumour, which is around 15-20 slides, will be digitised (approximately 250,000 whole slide images). This includes the index tumour slide that corresponds to the sequenced tissue, background tissue, and any immunohistochemical slides available. This is a unique feature of this dataset.
To control variation in image acquisition, the entire dataset will be scanned on a single scanning platform at a central NPIC scanning facility. The associated pathology reports will be curated from the original free-text formats to a structured, computer-readable format. This will be done using the Royal College of Pathologists minimum datasets for cancer and coded with SNOMED-CT and ICD-10 codes (Fig. 1), generating rich associated pathology metadata.
Conversely, radiology images are already digitised and in a standardised format known as DICOM. To enable movement of radiology images, we are using a system already used by hospitals to move images known as the Image Exchange Portal (IEP).
For security and governance, we anonymise the radiology images to allow for research use. To make use of the de-identified images, Genomics England have uniquely installed a Picture Archiving and Communication System (PACS).
Share your feedback
We believe in a human-centred approach to the products and services we provide to our users. If you would like to be part of user research to give feedback on our multimodal cancer research platform, share your views by completing this survey.
We'll continue to share updates on our platform as we digitise more data and build out our cloud-based analytics. You can also check out our other blogs to keep up with the work we do at Genomics England.