
The Genomics England Clinical Interpretation Partnership (GECIP) is a global community of researchers with access to our Research Environment.
We are immensely proud of what GECIP has achieved and the impact it has had on genomic research and healthcare. As the community continues to grow and the research outputs become more diverse, it is time for GECIP to evolve as well.
We are delighted to announce that starting from 19 September 2023, GECIP will begin to take on a new, streamlined, cross-cutting structure.
What does GECIP look like now?
Researchers in GECIP can access de-identified genomic and clinical data from the National Genomic Research Library (NGRL). They can use this data in their research to make scientific discoveries and gain patient-related insights.
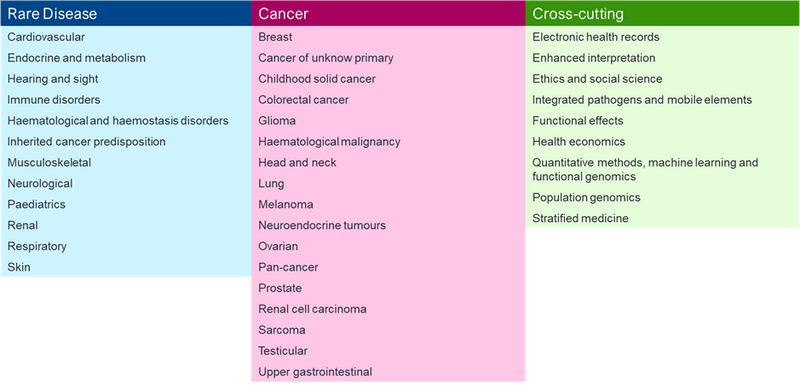
Currently, GECIP is split into 3 overarching categories: Rare disease, Cancer, and Cross-cutting. Together, these broad categories are composed of 38 subgroups called domains (see below). Each domain is a community of academic researchers and healthcare professionals investigating a specific disease or focus area.
Our thriving research community has had a wide range of successes so far, having produced over 165 publications, returning more than 2,000 diagnoses to NHS clinical teams, informing clinical practice, developing treatments, and shaping how genomic medicine benefits patients in the NHS.
With over 580 research projects currently active and more joining every day, we expect this activity and impact to continue increasing.
Why is GECIP changing?
The current GECIP structure was designed to support the 100,000 Genomes Project. However, over the last decade our goals and research community have evolved significantly.
We have undergone rapid changes in size and composition, and incorporated several impactful new initiatives such as Diverse Data, the Newborn Genomes Programme, Cancer 2.0, and the Genomic Medicine Service.
Our existing structure is not effectively designed to support this new research ecosystem.
The current structure of 38 domains has led to silos in which knowledge and skills are not being shared optimally within the community. This limits our ability to provide effective support to each domain, and can be restrictive of the research area and approach.
Our objective is to introduce a new structure, along with an updated leadership model and streamlined governance processes. It will optimally serve our evolving research community and maximise participant benefit. In particular, the new model prioritises:
- Simpler data access for approved individuals or projects, making it easier for GECIP members to conduct research.
- Allowing Genomics England to work with GECIP domains and their members to boost activity in prioritised research areas.
- Building a community active in knowledge sharing and research development, and identifying where support from Genomics England is having the most impact.
Streamlining governance should reduce the burden on domains and ease access to the NGRL.
Rather than domains having the responsibility to approve each individual’s NGRL access, the process will be managed by the Genomics England Access Review Committee.
We will continue to strongly support current GECIP activities. For example, maximising the proportion of participants who receive a diagnosis in the diagnostic discovery of rare disease.
What is the new structure?
The 38 disease-specific domains will be transformed into 8 new cross-functional communities.
Below are the 8 new complementary communities, along with the type of research they will encompass:
- Pan-cancer and molecular oncology
Research to find functional alterations and impaired biological mechanisms common to all/many cancers, and to establish whether there are common mutational signatures that affect treatment efficacy and toxicity.
- Predisposition and screening
Research encompassing use of genomics to identify undiagnosed patients with diseases or to predict those that are likely to develop diseases. This may include newborn screening research, polygenic risk scores, and familial cancer syndromes.
- Variant discovery and clinical interpretation
Diagnostic discovery and functional validation of researcher-identified diagnoses. General research identifying diagnostic endpoints.
- Bioinformatics and machine learning
Developing methods for analysing genomic data for research or clinical application. Providing bioinformatics support, and research using the NGRL dataset to develop algorithms and tools in relevant research and clinical areas.
- Therapeutic innovations and trials
Research encompassing pharmacogenomics, identification of potential treatment targets and therapeutic endpoints, and design and performance of genomics-informed clinical trials.
- Population genomics
Research that uses ancestry and population-level genomic data to study mutation and heritability patterns, (to fill in missing data for disease studies, and to analyse/improve NGRL data diversity).
- Implementation and data enhancement
Research in ethics/social science, health economics, electronic health records, functional activity for data federation, and improving clinical data/secondary health data.
- Genotype-phenotype association
Research that correlates phenotype and genotype at scale and developing and testing methods for doing so.
By having 8 cross-functional areas, this new model will ease communication and facilitate sharing of skills.
It will promote interaction whilst reducing areas of overlap and redundancy, helping to unlock the true potential of cross-cutting research, and could be transformative.
How did we make decisions?
The structure was designed to cover the core areas of Genomic England research as well as novel research, whilst encouraging more interaction and synergy between the communities.
We have developed this model over several years in consultation with researchers, domain leads, members of the GECIP Board, the Scientific Advisory Committee, and the internal Genomics England community.
We have worked with our stakeholders to identify blockers in the current model, and developed the new model to address these.
The overwhelming feedback was that we needed more synergistic communities, for which Genomics England could proactively and meaningfully drive interactions.
The new communities have been designed to unify research goals and meet previously unfulfilled needs. For example, the Variant discovery and clinical interpretation community addresses requests for a mechanism allowing researchers to share diagnostic findings and lessons in best practice.
How will changes affect research and researchers?
The new structure will make it easier to register new projects, collaborate with others, and receive increased support from Genomics England.
Ultimately, this will enable those using the Research Environment to maximise the value of their work for patients and participants.
Below are some important questions that users of the Research Environment might want to know...
How will these changes affect existing researchers and research projects?
For researchers and projects already registered with GECIP, their work will not be interrupted, and data access will remain the same. However, members will need to join one of the new communities and all projects will be realigned.
How will researchers receive more support from Genomics England?
With the number of GECIP communities decreasing from 38 to 8, there will be a lot more scope for Genomics England to provide strategic support.
This support will take the form of regular workshops, meetings, and training opportunities focused on specific but interdisciplinary goals.
There will also be more Genomics England initiatives to support members of the research community. For example, our teams from the Newborns, Cancer 2.0 and Diverse Data programmes will attend meetings to provide updates on new data, and discuss how we can best involve the research community.
Likewise, we will continue our recent work to help support early career researchers.
How will researchers know which new domain to join?
A detailed description of each domain will be released in the coming weeks to help researchers align themselves. Users of the Research Environment will have a say in deciding where their research sits best within the new structure.
How will my current GECIP network fit into this new structure?
Existing special interest groups will be free and encouraged to continue working together within the new structure. We want to retain existing networks that are effectively collaborating, and better link them up with Genomics England and other research groups.
Next steps...
The new communities will replace the existing cross-cutting domains starting mid-September. Then, over the following 6 months, the medical specialty-specific rare disease and cancer domains will be phased out, and the new communities will become the only GECIP communities in our research community.
More information (including detailed descriptions of each domain) will be released soon.
We look forward to these next steps with the research community, and to embark on this transformative journey together.
If you have any questions about the changes or exciting ideas for what we could achieve, please get in touch: [email protected].


