Cloud-based multimodal machine learning at Genomics England
By Tom Dyer, Sam Barnett, Andreia Rogerio and Golestan Karami onThe Genomics England Multimodal programme is collecting and analysing linked pathology, radiology, genomic and clinical data from patients with cancer. The aim being to improve patient-driven healthcare. Multimodal research brings unique challenges for researchers, such as extracting meaningful data representations, working in the cloud, and explaining complex model predictions. In this blog, we introduce several open-source codebases and applications to accelerate research on this dataset.
Multimodal research at Genomics England
As part of the Cancer 2.0 programme, Genomics England is assembling a world-leading multimodal dataset, bringing together linked genomic data, medical imaging and other clinical data. The project will deliver over 200,000 digitised pathology images and 400,000 radiology studies, associated with 17,000 cancer patients who were originally recruited as part of the 100,000 Genomes Project.
In the future, we aim to make this new data available to researchers in the Genomics England Research Environment, where whole genome sequences are already available. This will allow researchers to conduct multimodal analyses, with the potential to build a greater understanding of cancer progression, treatment response, and patient outcomes.
Machine learning and AI can supercharge this type of multimodal research, with the ability to combine high-resolution data types like digital pathology images and genomic sequences. Models that combine clinical, genomic, radiology and pathology information can make predictions about patient outcomes. These predictions try to mirror how treatment decisions are made by multi-disciplinary teams of experts in the NHS. Such technology therefore holds potential to deliver more personalised and effective care for patients with cancer in the future.
To support machine learning analysis, a robust cloud-based environment capable of handling large-scale data analysis is essential. We have been exploring these 2 components together to fully harness their potential.
In this blog, we will introduce the main challenges researchers face when working with multimodal datasets. We will showcase several solutions we’ve developed with machine learning in the Cloud at Genomics England to address these challenges and accelerate research.
The challenges of multimodal machine learning
Extracting meaningful data representations
Representation learning is a field of machine learning that involves processing unstructured data types (like images or text) and extracting numerical versions of the data. The representations, often referred to as embeddings or features, can be used for downstream machine learning tasks and easily combined with other data types. Read more in our previous blog about learning meaningful data representations for biomedical research.
Pipelines for representation extraction can be challenging to build as they require detailed knowledge of each data modality, and not every research team has genomic, radiology, and pathology experts available. Furthermore, for large data types like digital pathology scans, which can be several gigabytes in size, performing feature extraction can be both time consuming and costly. Making precomputed representations available can minimise the requirements for specialist domain expertise.
Working in the Cloud
Making such a large quantity of imaging and genomic data available securely to researchers at scale poses a grand infrastructure challenge. To tackle that, we are expanding our cloud-based Research Environment to allow users to access the data in a way that also safeguards patients, while still providing flexibility for machine learning development.
Many researchers are already familiar with cloud-based workflows, however, for some it represents a steep learning curve. Furthermore, any codebases that researchers want to use with our data must be brought into and adapted for a cloud-based environment.
We have developed several codebases that show how to get started in the cloud, and how to use Genomics England data from within. Our use-cases use AWS and Sagemaker – AWS machine learning service – to accelerate development.
Explaining model predictions
Many approaches to multimodal research involve not only training predictive models, but identifying the driving forces behind their predictions. This can help researchers identify image and genomic biomarkers that give us useful clues about patient prognosis, or predict response to treatments.
Extracting explanations can be complex, technical work, and often also requires expertise in specific clinical modalities (for example, highlighting a region of focus in a pathology image might still require an expert pathologist to explain the contents). Having prebuilt and model-agnostic explainability methods available can greatly reduce the amount of technical work required, and reduce the need for expert clinical team member time.
Prebuilt codebases to accelerate research
Over the past year, we have been working on internal solutions to many of these problems, enabling internal researchers to begin to extract value from the collected data. Now we are demonstrating the fruits of these efforts, so that when we can enable this work more widely researchers will be able to get a head start on their development. By sharing the resources detailed below, we hope to reduce the time users spend on mundane setup tasks and help them focus their efforts on scientific discovery.
Extracting meaningful data representations
Using pre-trained transformers to extract pathology features
Effective feature extraction from digital pathology images is a challenge of significant research focus. It is difficult not just because of the size of the high-resolution images, but also because relevant information can be found at different scales, with data from the cell, tissue and whole slide level contributing to diagnosis and treatment decisions.
Many open-source solutions are available to tackle this problem. These handle the loading of whole slide images, chunking of data into manageable regions, and passing them through machine learning models to generate usable representations. We are investigating solutions that can run in our cloud environment with the aim of making these available in the future, so that researchers with little experience in pathology can build powerful models.
We have adapted a leading pathology feature extraction method developed by the Mahmood lab at Harvard University, the Hierarchical Image Pyramid Transformer (HIPT) method and deployed it within AWS SageMaker. This method uses the powerful transformer architecture to extract features at three different magnification levels to give a final information rich embedding.
The features extracted by HIPT have been shown to encode relevant information that can be used for several downstream tasks, such as diagnosis and survival prediction, with published results showing state-of-the-art performance. The implementation in our environment makes use of GPU scaling offered by SageMaker Processing Jobs, and stores outputs in S3 buckets for downstream use.
This code can serve as a boilerplate example for researchers looking to deploy their own feature extraction methods, or they can access pre-extracted features to save time and compute cost in their research projects.
Genomic feature extraction with large language models
In recent years, the application of powerful Large Language Models (LLMs) to genomic sequences has shown great promise for extracting meaningful features from sparse genomic data. These features can be used to train predictive models and be combined with other modalities to gain a better understanding of cancer.
However, these powerful LLMs are expensive to train and deploy and require many GPUs, meaning the benefit of this new technology is limited to researchers with sufficient resources. To tackle this, we have developed a codebase which extracts genomic features in our cloud environment and makes them available for downstream research without the need to run the computationally expensive models every time.
This codebase allows users to select a target gene within a patient cohort and generate features using the Nucleotide Transformer model, developed by InstaDeep. The extracted features are numerical embeddings and are posted into S3 cloud storage for easy access to train models in downstream tasks.
This work is the foundation to build high-quality LLM-based genomic features and support exploration of downstream use-cases, as well as benchmarking predictive models from genomic sequences.
Working in the Cloud
Multimodal survival prediction from pathology and genomic data
To enable machine learning analysis, data needs to be processed in the cloud. This means we must adapt our existing local codebases to the cloud-based environment, to run model training and analysis pipelines. To reduce the technical overhead of this process, we have implemented several open-source codebases to run in our AWS environment. These serve as boilerplate for other adaptations and can also be used as a starting point for future research.
One example of this is PORPOISE, a model developed by the Mahmood lab and trained on public datasets to predict survival of patients with cancer using genomic and pathology data. Implementation of this model in the Genomics England Research Environment required adapting the codebase to access data inputs from S3 buckets, initiating GPU-powered training jobs in the cloud, and effectively storing and analysing results.
This codebase is an example of how to adapt model training code to run within AWS and SageMaker, submitting training containers to Elastic Container Repository (ECR), selecting GPU-powered instance types and analysing results in SageMaker notebooks.
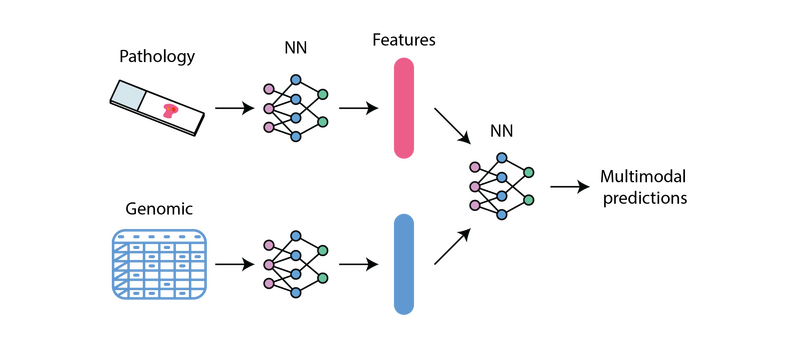
Representation of Multimodal Machine Learning achieved by the PORPOISE model from Chen et al.,
Lightweight machine learning workflows to enable rapid prototyping
One challenge of working with multimodal data is the sheer complexity of building code that can process several modalities and train predictive models. This can limit the potential exploration of researchers, as it is difficult to generate quick, proof-of-concept projects to test hypotheses.
To tackle this, we have developed a lightweight model training codebase called Tabular Trainers. This repository trains simple survival and classification models, taking pre-extracted features made readily available in S3 as inputs. The codebase follows a simple workflow, making use of several SageMaker native features, such as Training and Hyperparameter Optimisation Jobs, and can generate initial results in a matter of minutes.
The resulting codebase provides step-by-step Jupyter Notebooks to familiarise users with running experiments in the cloud, which can be customised to their cohort or task of interest. We have even seen that for many tasks, these simple models can achieve results comparable to that of published, complex deep learning models.
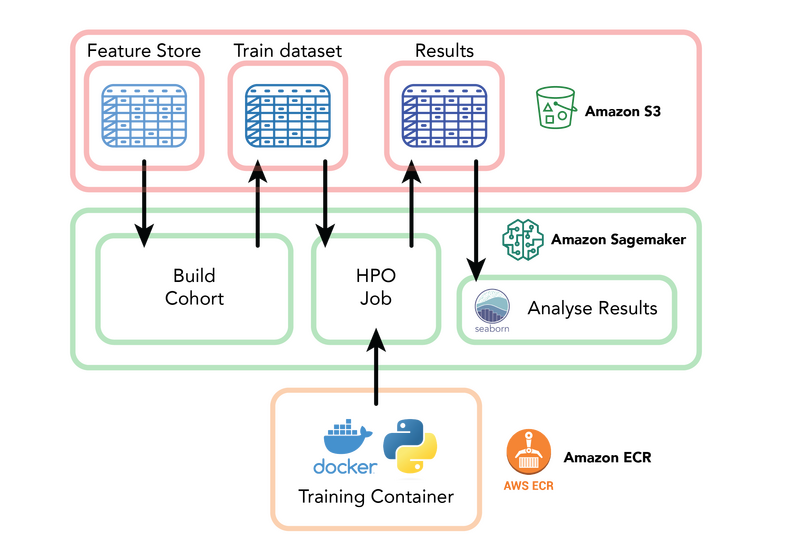
The Tabular Trainers repository uses AWS services including SageMaker, S3 and Elastic Container Repository to enable rapid machine learning prototyping.
Explaining model predictions
Model agnostic explanations for pathology images
Explaining the driving force behind model predictions can be one of the most challenging aspects of multimodal machine learning research. It can often require bespoke technical research and clinical expertise to interpret the output.
To assist in the generation of image-based explainability for pathology models, we have implemented a cluster-based Explainability Method inspired by work from researchers at Google Health to identify new colorectal cancer biomarkersolorectal cancer biomarkers. This method generates clusters of image patches which contain similar pathology image features, such as invasive cancer cells or regions of immune cells, such as lymphocytes.
The prevalence of these clustered features can then be compared to survival model predictions across a cohort, looking for cases where variation in survival predictions can be explained by a high or low level of a certain clustered feature.
This allows the generation of initial explanation of model predictions without having to invest technical time and speeding up understanding of model results. It also helps with identification of potential biases like image artefacts early in model lifecycles to minimise potential downstream impacts.
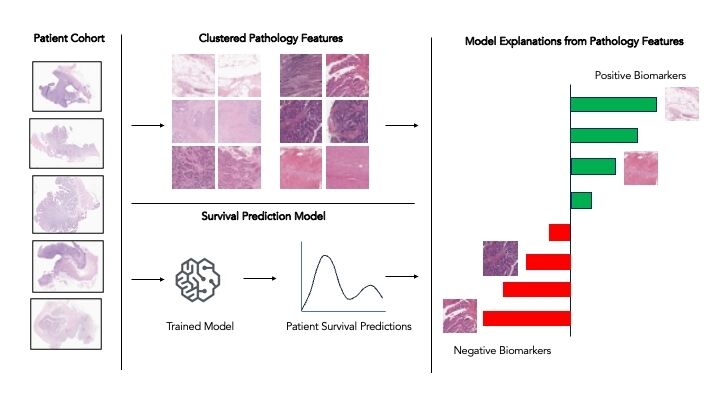
Representation of the workflow to cluster pathology features and generate cohort-level explanations of model predictions.
At Genomics England, we are continuing to work with research users to understand their challenges and improve their experiences within our Research Environment. Although at present these datasets are not widely available for external users, we are working on making it so in the future.
Acknowledgements
This work would not have been possible without the support of our colleagues Bojana Selinsek, Charlotte Jennings, Laura Valis, Cong Chen, Bohdan Kulynych, Nour Merzouki, Francisco Azuaje, Prabhu Arumugam, Bahareh Beshavardi, Luke Burton, Alistair Hall, Marko Cubric, Georgia Chan, Krishna Saha, and many others.
References:
- HIPT - https://github.com/mahmoodlab/HIPT?tab=readme-ov-file
- Nucleotide Transformer - https://www.biorxiv.org/content/10.1101/2023.01.11.523679v3
- PORPOISE - https://www.cell.com/cancer-cell/fulltext/S1535-6108(22)00317-8
- Google Health blog - https://research.google/blog/learning-from-deep-learning-a-case-study-of-feature-discovery-and-validation-in-pathology/


